Abstract
MotA and MotB are integral membrane proteins that form the stator complex of the proton-driven bacterial flagellar motor. The stator complex functions as a proton channel and couples proton flow with torque generation. The stator must be anchored to an appropriate place on the motor, and this is believed to occur through a putative peptidoglycan-binding (PGB) motif within the C-terminal periplasmic domain of MotB. In this study, we constructed and characterized an N-terminally truncated variant of Salmonella enterica serovar Typhimurium MotB consisting of residues 78 through 309 (MotBC). MotBC significantly inhibited the motility of wild-type cells when exported into the periplasm. Some point mutations in the PGB motif enhanced the motility inhibition, while an in-frame deletion variant, MotBC(Δ197-210), showed a significantly reduced inhibitory effect. Wild-type MotBC and its point mutant variants formed a stable homodimer, while the deletion variant was monomeric. A small amount of MotB was coisolated only with the secreted form of MotBC-His6 by Ni-nitrilotriacetic acid affinity chromatography, suggesting that the motility inhibition results from MotB-MotBC heterodimer formation in the periplasm. However, the monomeric mutant variant MotBC(Δ197-210) did not bind to MotB, suggesting that MotBC is directly involved in stator assembly. We propose that the MotBC dimer domain plays an important role in targeting and stable anchoring of the MotA/MotB complex to putative stator-binding sites of the motor.
Many bacteria swim by means of flagella, filamentous organelles that extend from the cell surface. A flagellum consists of at least three parts, the filament (helical propeller), the hook (universal joint), and the basal body (rotary motor) (27). The flagellar motor is fueled by the proton or sodium motive force across the cell membrane and can rotate both clockwise and counterclockwise (3, 20, 47). A recent high-resolution observation of flagellar motor rotation revealed a fine stepping motion of the motor rotation (39). The flagellar motor is an elaborate molecular nanomachine that converts electrochemical potential energy into mechanical work. The energy-coupling mechanism, however, is still not known.
Intensive genetic and biochemical studies of the flagellum have been conducted with Salmonella and Escherichia coli, and more than 50 gene products are known to be involved in flagellar assembly and function (26). Among them, only five proteins are responsible for torque generation. Three of them are the rotor proteins FliG, FliM, and FliN (45), which are mounted on the cytoplasmic face of the membrane-embedded MS ring made of FliF and form the C ring structure (15). The FliG/FliM/FliN complex is also called the “switch complex” because mutations in these proteins cause defects in switching between clockwise rotation and counterclockwise rotation in response to environmental conditions (45). Crystal structures have been reported for these rotor proteins (10, 11, 24, 30), and disulfide cross-linking experiments with structural information obtained from those crystal structures revealed subunit arrangements in the MS-C ring structure (25, 30, 32, 33). The other two proteins responsible for flagellar motor rotation are integral membrane proteins MotA and MotB (13, 40). MotA and MotB have four and single transmembrane segments, respectively (12, 49), and four copies of MotA and two copies of MotB form the stator complex (8, 22, 36, 37, 48), which functions as a proton channel to couple proton flux with motor rotation (4, 41). Each motor contains more than 10 MotA/MotB complexes around the MS-C ring (5, 7, 34). Conserved charged residues, Arg90 and Glu98, which are located in the cytoplasmic loop of E. coli MotA, interact with the conserved charged residues of the C-terminal domain of FliG (50), suggesting that these electrostatic interactions are important for torque generation. MotB has an absolutely conserved and functionally critical aspartic acid residue in its single transmembrane segment. This Asp (Asp32 in E. coli MotB) is believed to function as a proton-binding site in the channel for the motor function (51). Charge-neutralizing mutations of this residue cause a conformational change in the cytoplasmic domain of MotA containing Arg90 and Glu98 (21), providing a plausible hypothesis that protonation of this Asp residue may trigger a conformational change in the stator complex that acts on the rotor to drive its rotation.
MotB has a large periplasmic domain, which contains a putative peptidoglycan-binding (PGB) motif (14, 19) that is well conserved among proteins such as OmpA, Pal, and MotY, which are outer membrane proteins that interact with the peptidoglycan layer noncovalently. The PGB motif of MotB is believed to associate with the peptidoglycan layer to anchor the MotA/MotB stator complex around the rotor (12, 29, 44). However, the stators appear to be replaced frequently, even in the steadily rotating motor, as demonstrated by abrupt and stepwise drops and restorations of the rotation speed of the motor (5, 7, 39), which presumably reflects dynamic dissociation and association of the stator and the rotor. This suggests that the association of the PGB motif of MotB with the peptidoglycan layer is also highly dynamic. Hosking et al. (17) identified a segment of MotB that acts as a plug to prevent premature proton flow through the MotA/MotB complex and proposed that interaction of the MotA/MotB complex with the flagellar basal body could trigger both opening of the proton channel and unmasking of the PGB domain of MotB. Although high-resolution structural information about a few PGB proteins is now available (16, 31), little is known about how the stator is targeted to the rotor and how the PGB motif of MotB associates with the peptidoglycan layer near the basal body.
In this study, we have carried out a genetic and biochemical characterization of an N-terminally truncated Salmonella MotB fragment missing the N-terminal 77 residues (MotBC). We show here that MotBC forms a stable homodimer, as well as a small amount of a MotBC-MotB heterodimer and inhibits wild-type motility when exported to the periplasmic space and that this negative dominance effect is still retained albeit significantly reduced by a deletion variant of MotBC that stays a monomer. We discuss possible roles of MotBC in the assembly of the functional motor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and mutagenesis.
The bacterial strains and plasmids used in this study are listed in Table 1. To design N-terminally truncated fragments of Salmonella MotB, we used a web-based secondary structure prediction program, PSIPRED (18), and chose a surface-exposed, unstructured region for the N termini of the fragments. Mutations in the PGB motif of MotB were generated in plasmid pNSK7 or pNSK6 by the QuikChange 1-day site-directed mutagenesis method as described by Stratagene. In-frame deletions were generated as described by Toker et al. (43). DNA sequencing was done with an ABI PRISM 377 DNA sequencer (Applied Biosystems).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant property(ies) | Source or reference |
|---|---|---|
| E. coli strains | ||
| Novablue | Recipient for cloning experiments | Novagen |
| BL21(DE3) | Host for overexpression from the T7 promoter | Novagen |
| RP6894 | ΔmotAB | J. S. Parkinson |
| Salmonella sp. strain SJW1103 | Wild type for motility and chemotaxis | 46 |
| Plasmids | ||
| pTrc99A | Cloning vector | Pharmacia |
| pHMK11 | pTrc expression vector | This study |
| pACTrc | pTrc promoter, p15A replication origin, lacIq Cmr | G. M. Fraser |
| pET19b | T7 expression vector | Novagen |
| pET22b | T7 expression vector | Novagen |
| pHMK1609 | pHMK11/MotA+MotB-His8 | This study |
| pNSK6 | pET19b/MotBC-His6 | This study |
| pNSK7 | pHMK11/PelBL::MotBC-His6a | This study |
| pNSK8 | pTrc99A/MotBC-His6 | This study |
| pNSK11 | pET19b/MotBC | This study |
| pNSK28 | pHMK11/PelBL::MotBCa | This study |
| pNSK31 | pACTrc/MotA+MotB | This study |
| pNSK32 | pACTrc/MotA+MotB-His8 | This study |
| pNSK6-R218W | pET19b/MotBC(R218W)-His6 | This study |
| pNSK6-Δ(197-210) | pET19b/MotBC(Δ197-210)-His6 | This study |
| pNSK6-T197I/R218W | pET19b/MotBC(T197I/R218W)-His6 | This study |
| pNSK7-T197I | pHMK11/PelBL::MotBC(T197I)-His6a | This study |
| pNSK7-D198N | pHMK11/PelBL::MotBC(D198N)-His6a | This study |
| pNSK7-S215F | pHMK11/PelBL::MotBC(S215F)-His6a | This study |
| pNSK7-R218W | pHMK11/PelBL::MotBC(R218W)-His6a | This study |
| pNSK7-R223H | pHMK11/PelBL::MotBC(R223H)-His6a | This study |
| pNSK7-Δ(197-210) | pHMK11/PelBL::MotBC(Δ197-210)-His6a | This study |
| pNSK7-Δ(211-226) | pHMK11/PelBL::MotBC(Δ211-226)-His6a | This study |
| pNSK7-T197I/R218W | pHMK11/PelBL::MotBC(T197I/R218W)-His6a | This study |
A MotB fragment coding for residue 78 to C-terminal residue 309 (MotBC) was fused to a PelB leader sequence derived from pET22b at the N terminus and fused to His6 at the C terminus.
Preparation of whole-cell extract and a periplasmic fraction.
Cells were grown exponentially at 37°C in 5 ml LB medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) containing 100 μg/ml ampicillin. After addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), culture was continued at 37°C for another 1 h. The cells were harvested and resuspended in spheroplast buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.5 M sucrose) at a cell concentration equivalent to an optical density at 600 nm of 5. This cell suspension was diluted 10 times with water and used as the whole-cell sample. To prepare the periplasmic fraction, 100 μl of cell suspension was diluted five times in spheroplast buffer and incubated at room temperature for 20 min. After centrifugation (17,000 × g, 5 min), cells were carefully resuspended in 500 μl of 0.5 mM MgSO4 and placed on ice for 10 min. After centrifugation (17,000 × g, 5 min), the periplasmic fractions were collected.
Motility assays.
Swarming motility of SJW1103 transformed with appropriate plasmids was analyzed on TB soft-agar plates (1% [wt/vol] tryptone, 0.5% [wt/vol] NaCl, 0.28% [wt/vol] Bacto agar) containing 100 μg/ml ampicillin at 30°C typically for 6 h. IPTG was added as needed to a final concentration of 1 mM or 0.1 mM. To measure swimming speed, the cells were cultured in TB (1% [wt/vol] tryptone, 0.5% [wt/vol] NaCl) at 30°C until log phase. If necessary, IPTG was added at the beginning of the culture or at the log phase to a final concentration of 0.1 mM or 1 mM. The culture media were then diluted 1:10 in fresh TB to observe the motility of the cells under a phase-contrast microscope. The swimming speed of the cells was measured as described previously (2).
Purification of MotB fragments.
E. coli BL21(DE3) cells transformed with pNSK6 were collected by centrifugation and resuspended in buffer A (20 mM Tris-HCl [pH 8.0], 100 mM NaCl) containing one tablet of Complete protease inhibitor cocktail (Roche Diagnostics). The cells were disrupted, the soluble fraction was isolated by ultracentrifugation (186,000 × g, 30 min), and then supernatants were collected and loaded onto a set of tandem HiTrapSP (GE Healthcare), HiTrapQ (GE Healthcare), and HisTrap (GE Healthcare) columns connected in that order. MotBC-His6, which flowed through the HiTrapSP and HiTrapQ column but bound to HisTrap column, was eluted by a linear gradient of imidazole, collected, and further purified by a Sephacryl S-300 size exclusion column (GE Healthcare). Mutant variants of MotBC-His6 were purified in the same way. For purification of MotBC fragments without a His tag, we used plasmid pNSK11. The soluble fraction of BL21(DE3) carrying pNSK11 was loaded to the HiTrapSP-HiTrapQ columns connected in that order and equilibrated with buffer B. Flowthrough fractions containing MotBC were collected by adding (NH4)2SO4 to 45% saturation. The pellet was suspended in 10 ml of buffer B, and then MotBC was further purified by the Sephacryl S-300 size exclusion column as described above.
Analytical size exclusion column chromatography.
Analytical size exclusion chromatography was performed with a Superdex 75 HR 10/30 column (GE Healthcare) connected to an AKTA system (GE Healthcare). The column was equilibrated with buffer B and run at a flow rate of 0.7 ml/min. Bovine serum albumin (BSA) (67 kDa), ovalbumin (44 kDa), and chymotrypsinogen (25 kDa) were used for size markers.
Analytical ultracentrifugation.
Sedimentation equilibrium analytical ultracentrifugation was carried out with a Beckman Optima XL-A analytical ultracentrifuge with an AnTi 60 rotor as described previously (28). The purified samples of MotBC and MotBC(Δ197-210)-His6 were dialyzed against 20 mM Tris-HCl (pH 8.0) buffer solutions containing 100 mM and 300 mM NaCl, respectively, which were also used as the blank. Measurements were done at 20°C at 20,000, 22,000, and 24,000 rpm on the MotBC fragment and at 24,000, 26,000, and 28,000 rpm on MotBC(Δ197-210)-His6 with charcoal-filled Epon and quartz windows. Concentration profiles of the samples were monitored by absorbance at a wavelength of 280 nm and recorded at a spacing of 0.001 cm in the step mode, with 20 averages per step, for 10, 16, and 22 h after each rotor speed was reached. Equilibrium data were analyzed with the Beckman Optima XL-A/XL-I data analysis software, version 4.0, provided as an add-on to Origin version 4.1 (MicroCal Inc.). A global, single-species fit over two different loading absorbance (0.2 and 0.3 at 280 nm) and three rotor speeds, as described above, were calculated. The partial specific volumes, 0.730 ml/g for MotBC and 0.731 ml/g for MotBC(Δ197-210)-His6, used for analysis were based on the amino acid compositions of the proteins.
Antibodies and immunoblotting.
Purified MotBC-His6 was used to raise an anti-MotB antibody in rabbits (MBL Co., Ltd.). After the proteins in each fraction were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, immunoblotting with the polyclonal anti-MotB and anti-Pal antibodies was carried out as described previously (21). Detection was performed by the SuperSignal West Pico chemiluminescence procedure (Pierce).
Pull-down assay.
E. coli motA-motB deletion strain RP6894 was transformed with two plasmids, one encoding His-tagged MotBC with or without the PelB leader sequence at their N termini (pNSK7 or pNSK8) and the other encoding both nontagged MotA and MotB (pNSK31). A monomeric mutant variant, MotBC(Δ197-210), was also expressed from pNSK7-Δ(197-210) in place of MotBC. For the opposite His tag combination, we used plasmids pNSK28 (nontagged MotBC) and pNSK32 (MotA/MotB-His8). Overnight culture was inoculated into 10 ml fresh LB containing 100 μg/ml ampicillin and 25 μg/ml chloramphenicol and incubated at 37°C until log phase. After the addition of 0.1 mM IPTG, incubation was continued at 37°C for another 1 h. The cells were harvested and resuspended in 1 ml of spheroplast buffer, sonicated, and centrifuged (3,000 × g, 5 min). After centrifugation (16,000 × g, 15 min), the membrane fraction was suspended in 1 ml of buffer C (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM imidazole, 10% [vol/vol] glycerol), and then a detergent, dodecylphosphocholine (DPC; Anatrace Inc.), was added to a final concentration of 0.05% (wt/vol). After gentle shaking for 20 min at 30°C, insoluble materials were removed by centrifugation (16,000 × g, 15 min). The soluble fraction was mixed with 50 μl of Ni-nitrilotriacetic acid (NTA) agarose resin (Qiagen) prewashed with buffer C containing 0.05% (wt/vol) DPC. After gentle mixing at 30°C for 20 min, the unbound materials were removed by brief centrifugation (ca. 5 s). The resin was washed twice with buffer C with 0.03% (wt/vol) DPC (1 ml/wash) and then three times with buffer C containing 60 mM imidazole and 0.03% (wt/vol) DPC (1 ml/wash). After incubation for 1 min at room temperature, proteins were eluted with 100 μl buffer C containing 500 mM imidazole and 0.03% (wt/vol) DPC. Eluted materials were mixed with SDS loading buffer and boiled.
RESULTS
Multicopy effect of MotBC on motility.
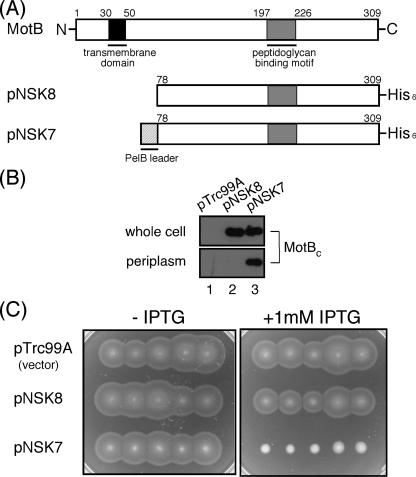
We constructed several N-terminally truncated variants of MotB that lack the transmembrane segment and placed them under the control of the IPTG-inducible trc promoter. These MotB fragments were fused to a PelB leader sequence (PelBL) at the N termini to direct their location to the periplasmic space. A His6 tag was also attached to these fragments at the C terminus to facilitate protein purification (Fig. 1A). We transformed wild-type Salmonella strain SJW1103 with these constructs, prepared the whole-cell and periplasmic fractions from the resulting transformants, and analyzed them by immunoblotting with polyclonal anti-MotB antibody (Fig. 1B). One of these constructs, consisting of residues 78 through 309 (PelBL-MotBC-His6) (expressed from pNSK7), was stably expressed and detected in the periplasmic fraction (Fig. 1B, lane 3). In contrast, MotBC-His6, which does not have the PelB signal sequence at its N terminus, was not detected in the periplasm (Fig. 1B, lane 2).
FIG. 1.
Multicopy effect of MotBC on the motility of wild-type cells. (A) Primary structures of the Salmonella MotB protein and an N-terminally truncated fragment missing the N-terminal 77 residues, MotBC. MotB has a single transmembrane domain (black) in the N-terminal region and a putative PGB motif (gray) in the large periplasmic domain. Plasmid pNSK7 encodes the MotBC fragment (residue 78 to C-terminal position 309) fused to a PelB leader sequence (PelBL, 22 amino acids, hatched) and a His6 tag at its N and C termini, respectively. Plasmid pNSK8 does not contain PelBL at the N terminus. (B) Periplasmic localization of the MotBC-His6 fragment. Immunoblotting with the polyclonal anti-MotB antibody of whole-cell proteins (whole cell) and periplasmic fractions (periplasm) prepared from SJW1103 transformed with pTrc99A (vector control), pNSK8, or pNSK7. (C) Swarming motility assay of SJW1103 carrying pTrc99A, pNSK8, or pNSK7 on soft-agar plates with or without 1 mM IPTG. Plates were incubated at 30°C for 6 h.
Expression of PelBL-MotBC-His6 with 1 mM IPTG resulted in a severely impaired swarming motility of wild-type cells on soft-agar plates, while expression of MotBC-His6 did not show any notable effect on motility (Fig. 1C), indicating that the periplasmic location of MotBC is critical for motility inhibition. The results were essentially the same when the IPTG concentration in the plate was reduced to 0.1 mM. Therefore, we used 0.1 mM IPTG for motility assays thereafter.
To investigate how flagellar motor rotation is affected by the periplasmic location of MotBC-His6, we measured the swimming speed of SJW1103 carrying pNSK7 or pNSK8 cultured to the log phase in the presence of 0.1 mM IPTG. The swimming speed of SJW1103 carrying pTrc99A or pNSK8 was not affected by IPTG induction (about 27 μm/s for both strains). In contrast, the swimming speed of SJW1103 carrying pNSK7 was 18 μm/s and 11 μm/s in the absence and presence of IPTG, respectively. Thus, the swimming speed was reduced significantly (to 40% of that of the wild type) by the addition of IPTG. Even in the absence of IPTG, the speed was reduced to about 60% of the control strains, probably because of the leakiness of the MotBC expression from plasmid pNSK7.
Multicopy effect of mutant variants of MotBC-His6 on motility.
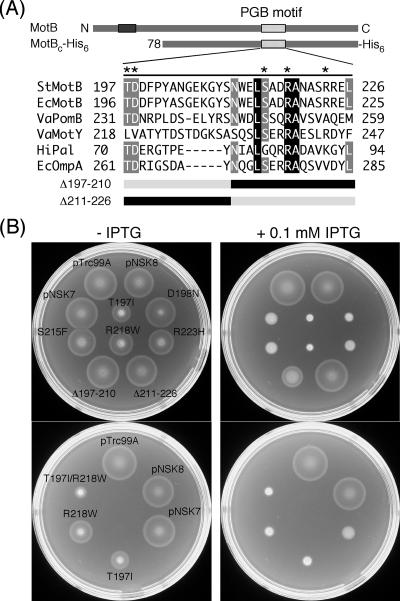
Blair et al. (6) have identified many point mutations in E. coli motB that give a Mot− (paralyzed flagella) phenotype. Most of them are located within the periplasmic domain of MotB, including the putative PGB motif (Fig. 2A) (14, 19, 29). These mutant variants also exhibit a negative dominance effect on the motility of the wild type, probably reflecting the displacement of functional MotB by a nonfunctional one. To investigate if these dominant-negative mutations affect the multicopy effect of MotBC on motility, we introduced five of these point mutations lying in the PGB motif into MotBC-His6. We also constructed two in-frame deletion variants, MotBC(Δ197-210) missing residues 197 to 210 and MotBC(Δ211-226) lacking residues 211 to 226 (Fig. 2A). These mutant variants were fused to the PelB signal sequence at their N termini. The plasmids containing these mutations were introduced into SJW1103, and the level of negative dominance on motility by these mutant versions of MotBC-His6 was assayed on soft-agar plates with or without 0.1 mM IPTG (Fig. 2B, upper two plates). All of the five point mutant variants still exhibited negative dominance. The D198N, S215F, and R223H mutant variants inhibited motility almost at the wild-type MotBC level (see pNSK7). The T197I and R218W mutants impaired motility more strongly than the wild-type MotBC fragment, as also shown even in the absence of IPTG (Fig. 2B, upper left plate). The (Δ197-210) deletion mildly inhibited the motility, but this dominance effect was much weaker than that of wild-type MotBC. The (Δ211-226) deletion did not exert any inhibitory effect (Fig. 2B, upper right plate).
FIG. 2.
Dominance properties of various mutant variants of MotBC-His6. (A) Multiple-sequence alignment of bacterial proteins containing a PGB motif and mutations generated in MotBC-His6. Sequences aligned are as follows: StMotB, Salmonella enterica serovar Typhimurium MotB; EcMotB, Escherichia coli MotB; VaPomB, Vibrio alginolyticus PomB; VaMotY, Vibrio alginolyticus MotY; HiPal, Haemophilus influenzae Pal; EcOmpA, Escherichia coli OmpA. The multiple-sequence alignment was done by the ClustalW software (42). Asterisks indicate residues mutated in this study. Residues shown in the black (or gray) box with white letters are completely conserved (or well conserved) among these six proteins. The point mutations are those originally reported by Blair et al. (6) to produce a dominant-negative Mot− phenotype in E. coli. Δ197-210 and Δ211-226 are deletion mutants that lack residues 197 to 210 and 211 to 226, respectively. (B) Swarming motility assay of wild-type strain SJW1103 transformed with plasmids pTrc99A (vector control), pNSK8 (MotBC-His6), pNSK7 (PelBL-MotBC-His6), T197I [PelBL-MotBC(T197I)-His6], D198N [PelBL-MotBC(D198N)-His6], S215F [PelBL-MotBC(S215F)-His6], R218W [PelBL-MotBC(R218W)-His6], R223H [PelBL-MotBC(R223H)-His6], Δ197-210 [PelBL-MotBC(Δ197-210)-His6), Δ211-226 [PelBL-MotBC(Δ211-226)-His6), and T197I/R218W [PelBL-MotBC(T197I/R218W)-His6]. Cells were inoculated onto the same positions of soft-agar plates with or without 0.1 mM IPTG. The strain names are indicated only on the left plate. Plates were incubated at 30°C for 6 h.
We next examined whether the T197I and R218W mutations show an additive effect on motility inhibition (Fig. 2B, lower two plates). The T197I/R218W double-mutant variant exhibited much stronger negative dominance on motility of wild-type cells than either of the single-mutant variants, as clearly shown in the absence of IPTG (Fig. 2B, lower left plate).
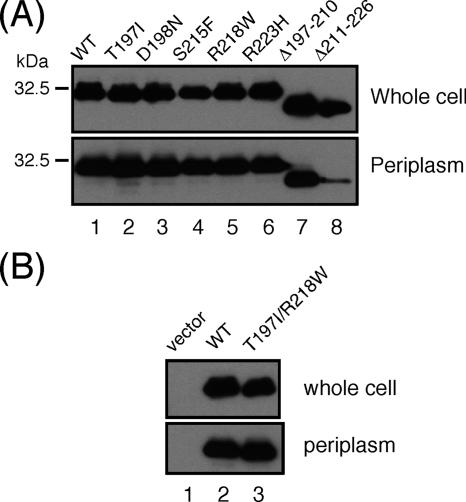
To examine the level of protein expression and periplasmic localization of these mutants, we prepared the whole-cell and periplasmic fractions from SJW1103 carrying the plasmids and carried out immunoblotting with the anti-MotB antibody (Fig. 3). All of the point mutant variants, the (Δ197-210) deletion, and the double-mutation variant were expressed and exported into the periplasm at wild-type levels. However, only a small amount of the (Δ211-226) deletion was detected in the periplasm although it was expressed at a wild-type level (Fig. 3A, lane 8). Therefore, the absence of an inhibitory effect on motility by this deletion fragment was probably due to the defect in its periplasmic localization.
FIG. 3.
Periplasmic localization of mutant variants of MotBC. (A) MotBC with single mutations or deletions. (B) MotBC with a T197I/R218W double mutation. Immunoblotting with the anti-MotB antibody of whole-cell proteins and periplasmic fractions of SJW1103 transformed with the wild type (WT), pNSK7; T197I, pNSK7(T197I); D198N, pNSK7(D198N); S215F, pNSK7(S215F); R218W, pNSK7(R218W); R223H, pNSK7(R223H); Δ197-210, pNSK7(Δ197-210); Δ211-226, pNSK7(Δ211-226); and T197I/R218W, pNSK7(T197I/R218W).
Dimerization of MotBC.
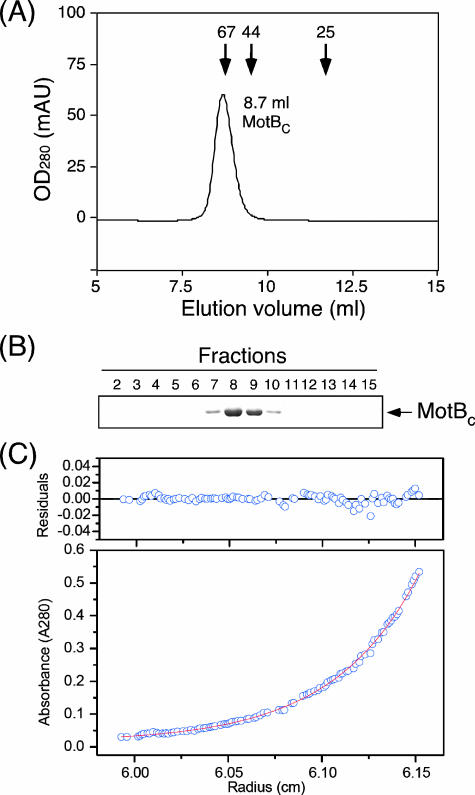
To investigate the oligomerization property of MotBC, we carried out sedimentation equilibrium analytical ultracentrifugation and analytical size exclusion chromatography. We constructed plasmid pNSK11, which overproduces MotBC without the PelB leader sequence and the His6 tag, to avoid complexity in interpreting the results of these measurements (35). Untagged MotBC was purified and subjected to analytical size exclusion column chromatography with a Superdex 75 HR 10/30 column (Fig. 4A and B). MotBC eluted from the column at a volume of 8.7 ml, which is close to the elution position of BSA (67 kDa), indicating that the size of MotBC in solution is much larger than the deduced size of a MotBC monomer (25.7 kDa) and suggesting that MotBC forms an oligomer in solution. To measure the molecular size of the oligomer more precisely, we performed sedimentation equilibrium analytical ultracentrifugation, which can determine the molecular mass of particles in solution independently of their shape. The same batch of the MotBC sample was used for the measurements at three different protein concentrations, and the results were basically the same. The manufacturer's software was used to test several models to fit the profiles obtained, and a single-species model produced the best fit in terms of low residuals (Fig. 4C). The calculated molecular mass was 50.6 kDa, which corresponds almost exactly to that of the dimer of MotBC.
FIG. 4.
Hydrodynamic properties of MotBC protein. (A) Elution profile of purified MotBC by analytical size exclusion chromatography with a Superdex 75 HR 10/30 column. Arrows indicate the elution peaks of the marker proteins BSA (67 kDa), ovalbumin (44 kDa), and chymotrypsinogen (25 kDa) at 8.8 ml, 9.5 ml, and 11.7 ml, respectively. OD280, optical density at 280 nm; mAU, milliabsorbance units. (B) SDS-polyacrylamide gel electrophoresis of elution fractions from panel A. Fractions 7 to 10 correspond to the volume from around 7.5 to 10 ml. (C) Sedimentation equilibrium analytical ultracentrifugation of MotBC. Open circles are data points, and the continuous line is a model fit. A concentration profile of MotBC (initial absorbance at 280 nm of 0.2, measured at 24,000 rpm) is shown at the bottom. For data fitting, we performed a global fit to six data sets from two different protein concentrations and three rotor speeds. The residuals due to deviation of the data from this line are shown at the top. The molecular mass obtained was 50.6 kDa, indicating that MotBC is a dimer. Measurements were done at room temperature.
To test if the C-terminal His tag affects the dimer formation of MotBC, we purified MotBC-His6 from the periplasmic fraction of wild-type cells carrying pNSK7 by HisTrap affinity chromatography and ran it on a Superdex 75 HR 10/30 column. MotBC-His6 was eluted at a volume of 9.0 ml from the column (data not shown), indicating that MotBC-His6 in periplasm, which exerts an inhibitory effect on flagellar motor rotation, also forms a dimer.
Dimerization of mutant MotBC-His6 fragments.
To examine if the mutations within the PGB motif affect the dimerization of MotBC, we purified His-tagged versions of these mutant fragments and analyzed them by analytical gel filtration chromatography with a Superdex 75 HR 10/30 column (Fig. 5A).
FIG. 5.
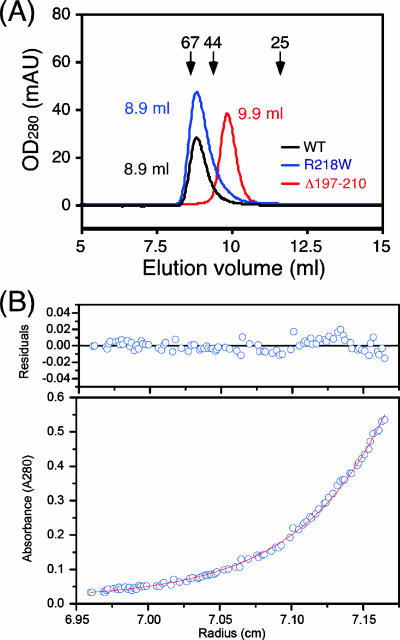
Hydrodynamic properties of MotBC mutant proteins. (A) Elution profiles of purified MotBC-His6 fragments determined by analytical size exclusion chromatography with a Superdex 75 HR 10/30 column. Black line, wild-type (WT) MotBC (peak at 8.9 ml); blue line, MotBC(R218W) mutant protein (peak at 8.9 ml); red line, MotBC(Δ197-210) mutant protein (peak at 9.9 ml). Arrows indicate the elution peaks of the size marker proteins BSA (67 kDa), ovalbumin (44 kDa), and chymotrypsinogen (25 kDa) at 8.6 ml, 9.4 ml, 11.6 ml, respectively. OD280, optical density at 280 nm; mAU, milliabsorbance units. (B) Sedimentation equilibrium analytical ultracentrifugation of MotBC(Δ197-210)-His6. Open circles are data points, and the continuous line is a model fit. A concentration profile for MotBC(Δ197-210)-His6 (initial absorbance at 280 nm of 0.2, measured at 28,000 rpm) is shown at the bottom. For data fitting, we performed a global fit to six data sets from two different protein concentrations and three rotor speeds. The residuals due to deviation of the data from this line are shown at the top. The molecular mass obtained was 24.0 kDa, indicating that MotBC(Δ197-210)-His6 is a monomer. Measurements were done at room temperature.
Purified MotBC(R218W) eluted at the same volume as the wild type (8.9 ml), indicating that the size and shape of the R218W mutant variant are essentially the same as those of wild-type MotBC. However, MotBC(Δ197-210) eluted at a volume of 9.9 ml. We performed sedimentation equilibrium analytical ultracentrifugation to precisely determine the molecular mass of the particle that this deletion variant forms in solution (Fig. 5B). The model fitting of the obtained profiles gave a molecular mass of 24.0 kDa, which is close to the deduced molecular mass of a MotBC monomer (25.0 kDa).
We also purified MotBC(T197I/R218W) and analyzed its molecular size by analytical gel filtration chromatography. This double-mutant protein was eluted at a volume of 9.0 ml (data not shown), indicating that this also forms a dimer in solution.
These results show a clear positive correlation between the dimerization of MotBC fragments and the level of motility inhibition. Wild-type MotBC, MotBC(R218W), and MotBC(T197I/R218W), which form stable dimers, all impaired motility, whereas MotBC(Δ197-210), which is monomeric, showed a reduced inhibition effect, although the level of inhibition was still significant.
Association of MotBC with MotB.
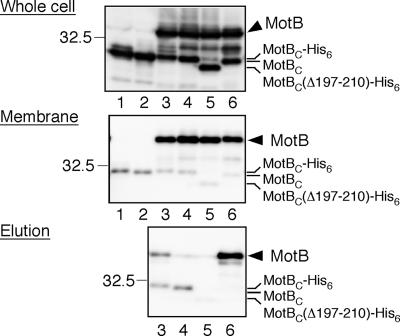
MotBC forms a dimer in solution, but full-length MotB is also known to form a homodimer in the stator complex (8, 9), raising the possibility that the multicopy inhibitory effect of MotBC on motility may be caused by titration of the endogenous full-length MotB protein by the plasmid-borne MotBC fragment in the periplasm, interfering with the formation of the functional MotA/MotB complex to be installed in the motor. To investigate this possibility, we analyzed the association of MotBC with MotB by pull-down assays (Fig. 6). If MotB associates with MotBC exported into the periplasm, it should be copurified with a His-tagged variant of MotBC by Ni-NTA affinity chromatography. MotBC-His6 with or without the PelB leader sequence was coexpressed with MotA and MotB in the E. coli ΔmotA-motB double-null mutant. Both MotBC and MotB were expressed at similar levels (Fig. 6, top panel, lanes 3 and 4). Although most of MotBC was in the soluble fraction (data not shown), small amounts of MotBC were found in the insoluble membrane fractions, at almost the same level for nonsecreted and secreted variants of MotBC (Fig. 6, middle panel, lanes 3 and 4). These membranes were solubilized with DPC and then mixed with Ni-NTA resin. After the resin was washed, MotBC-His6 was eluted by a high concentration of imidazole (Fig. 6, bottom panel, lanes 3 and 4). A significant amount of MotB was coisolated with MotBC-His6 exported to the periplasm (lane 3), but little was coisolated with that in the cytoplasm (lane 4). We also tested the binding of MotBC(Δ197-210), a monomeric variant of MotBC, to MotB. MotBC(Δ197-210)-His6 was detected in the membrane fraction, but only a small amount of MotB was coisolated (Fig. 6, bottom panel, lane 5).
FIG. 6.
Coisolation assay of MotBC variants with MotB. MotBC-His6 designed to be exported into the periplasm (expressed from plasmid pNSK7, lane 1) and expressed in the cytoplasm (pNSK8, lane 2) was coexpressed with MotA and MotB (expressed from compatible plasmid pNSK31) (lanes 3 and 4). The expression levels of MotB and MotBC were similar in both cases. The membranes were then isolated, solubilized with the detergent DPC, and mixed with Ni-NTA resin. MotBC-His6 and its associated proteins were eluted by imidazole, and samples prepared from each step were analyzed by immunoblotting with anti-MotBC antibody. A monomeric variant of MotBC [MotBC(Δ197-210) expressed from pNSK7-Δ(197-210)] was also examined in the same way (lane 5). An alternative combination (MotA/MotB-His8 and tagless MotBC expressed from pNSK32 and pNSK28, respectively) was also examined (lane 6). Top panel, whole-cell extracts; middle panel, membrane fraction; bottom panel, eluted products. The 32.5-kDa marker position is shown at the left of each panel.
Alternatively, the His8 tag was attached to MotB instead of MotBC, and then pull-down assays were carried out. Again, MotBC was found in the membrane fraction but no MotBC was coisolated with MotB-His8 (Fig. 6, bottom panel, lane 6).
These results suggest that MotBC can associate with MotB in the periplasm, probably contributing to the motility inhibition to some extent. However, the amount of the MotBC-MotB heterodimer is much smaller than that of the MotBC homodimer, as shown in Fig. 6. Note that there is always some nonspecific association of MotBC with the membrane, even in the absence of MotA and MotB, regardless of whether or not MotBC was exported to the periplasm (Fig. 6, middle panel, lanes 1 and 2). The faint bands of MotB coisolated with MotB fragments by Ni-NTA affinity chromatography are also likely to be the results of nonspecific binding of MotB to the resin.
DISCUSSION
Asai et al. have reported that a chimeric stator complex consisting of PomA and a chimeric PotB protein, a fusion of the N-terminal transmembrane segment of V. alginolyticus PomB and the C-terminal periplasmic segment of E. coli MotB, is functional in the E. coli flagellar motor, while the wild-type PomA/PomB complex is not (1), suggesting that an appropriate periplasmic domain of the stator complex is required for the association of stators with the basal body to form a functional motor. In this study, to investigate the roles and mechanisms of the periplasmic domain of MotB for the stator assembly, we have analyzed the C-terminal periplasmic domain of MotB (MotBC) and obtained evidence that MotBC forms a stable dimer in the periplasm and inhibits flagellar motor rotation.
MotBC, consisting of residues 78 through 309, inhibited the swarming motility of wild-type cells on soft-agar plates when it was exported to the periplasmic space (Fig. 1). In agreement with this, the swimming speed of wild-type cells expressing and exporting MotBC to the periplasm was significantly reduced. The T197I and R218W variants exhibited stronger negative dominance effects than wild-type MotBC, while MotBC(Δ197-210), an in-frame deletion mutant in the PGB motif, exhibited a reduced dominance effect (Fig. 2B). Wild-type MotBC and the point mutant variants of MotBC formed a stable homodimer, while the deletion variant was monomeric (Fig. 4 and 5), suggesting that dimerization of MotBC strengthens the negative dominance effect.
It has been estimated that there are at least 11 copies of the stator complex around the rotor of the flagellar motor (34). A decrease in the number of functional stators in the motor slows down the rotation speed (39). Therefore, the motility inhibition caused by overexpressed MotBC in the periplasm is likely to be due to a decrease in the number of functional stators. The question is how MotBC interferes with the assembly of functional stators into the motor. There are three possibilities. (i) MotBC has a motif to bind to the stator-binding sites of the flagellar basal body and occupies them, strongly in the dimer form and weakly in the monomer form; (ii) MotBC forms a heterodimer with endogenous MotB and thereby interferes with the formation of functional stators; or (iii) MotBC dimers compete with endogenous MotB for binding to MotA and thereby interfere with the formation of functional stators.
The results of the coisolation experiments with His-tagged MotBC and full-length MotB (Fig. 6) supports possibility 2. A small but significant amount of MotBC-His6 was found in the membrane fraction and copurified with full-length MotB by Ni-NTA affinity chromatography only when MotBC-His6 was exported into the periplasm. Since the motility inhibition was seen only when MotBC was located in the periplasm, the MotB-MotBC interaction in the periplasm might be responsible for the motility inhibition seen.
The amount of the MotB-MotBC heterodimer, however, is quite small compared to that of the MotBC homodimer (Fig. 6). The majority of MotBC forms a stable homodimer (Fig. 4 and 5). The stability of the MotBC homodimer is also supported by our nuclear magnetic resonance measurements of MotBC at three different temperatures (30°C, 40°C, and 50°C) (Y. Sudo and C. Kojima, personal communication). Moreover, even a monomeric variant of MotBC shows inhibition, albeit relatively weak, of motility. These results support possibility 1. The MotBC fragment might actually contain the targeting signal to drive the installation of the stator complex into the flagellar motor. To test this, in vivo imaging of MotBC behavior with a fluorescent protein such as “mCherry” (38) would be a good approach.
Although dominant-negative mot mutations within the putative PGB motif (6) seem to interfere with the association of the PGB motif with peptidoglycan, these mutations enhanced the motility inhibition by MotBC (Fig. 2). These apparently conflicting results cannot simply be explained by possibility 1 because the number of MotBC dimers or monomers occupying the stator-binding sites would be decreased if the binding of the PGB motif with peptidoglycan was weakened. However, since no structural data are available for the interaction between the PGB motif and peptidoglycan, it would also be possible that the mutations strengthened the binding in the case of MotBC, which has much more positional and orientational freedom than MotB in binding to the basal body. More-detailed analyses are required.
MotA was not detected in the elution fraction of the MotB-MotBC heterodimer by immunoblotting with the polyclonal anti-MotA antibody (data not shown), suggesting that the MotB-MotBC heterodimer may not form a complex with MotA. Alternatively, MotA may be dissociated from the MotB/MotBC heterodimer by stringent washes during purification. To test these possibilities, we need to examine whether MotA could be coisolated with MotBC-His6 in the absence of MotB.
It has been reported that MotB forms a dimer at its single transmembrane segment (9). In this study, we showed that the periplasmic domain of MotB alone suffices to form a stable dimer in the periplasm. Taken together, two MotB molecules are likely to associate with each other along their entire lengths. The MotBC dimer domain of the MotA4/MotB2 complex may play an important role in targeting the complex to its binding site and anchoring it to the motor to be the stator. In the stator resurrection experiments (5, 7, 39), abrupt drops in the rotation rate were observed rather frequently, and this may reflect dissociation or turnover of stators from the motor. In fact, a recent study by the fluorescence photobleaching technique has shown a turnover of green fluorescent protein-fused MotB between the membrane pool and the motor (23), suggesting that the interactions between the MotA/MotB complex and its target site on the motor are dynamic and that MotB does not always associate with peptidoglycan even though MotB has a highly conserved PGB motif. Our preliminary PGB assay showed that neither MotB nor MotBC was found in the PG-associated fraction, while Pal, a PGB protein, showed a strong association with peptidoglycan (data not shown), suggesting that the PGB site of MotB is opened and activated only upon binding of the MotBC dimer domain to the basal body. So far, there have been no reports showing evidence for specific stator-binding sites on and around the basal body. The crystal structure of the C-terminal domain of RmpM (RmpM_Cter) indicates that RmpM_Cter may exist as a dimer with two putative PGB sites located at sites opposite each other, suggesting that an RmpM dimer could simultaneously bind to two glycan chains (16). The MotBC dimer domain may bind to peptidoglycan in a similar manner, since remarkable structural similarities are found among PGB proteins (16, 31). However, as the association of the MotA/MotB complex with the flagellar motor is highly dynamic, the association of MotB with peptidoglycan would presumably be more transient and dynamic than that of outer membrane proteins such as RmpM. Therefore, it would be quite interesting to see how the MotBC dimer domain of the MotA/MotB complex behaves in vivo and how it associates with the peptidoglycan layer after the MotA/MotB complex is installed into the motor. To address these questions, MotBC would be a useful tool. Further efforts to understand the mechanism of motility inhibition by MotBC and to establish a PG association assay are ongoing, together with crystallization screening of MotBC to obtain structural insight into the stator-anchoring mechanism.
Acknowledgments
We thank Michio Homma and David Blair for critically reading the manuscript and stimulating discussion. We acknowledge Sandy Parkinson for a gift of the strain RP6894, Gillian Fraser for a gift of the pACTrc vector, Hajime Tokuda for a gift of the polyclonal anti-Pal antibody, Sachi Tatematsu for technical assistance, and Yuki Sudo and Chojiro Kojima for communicating unpublished results. We also thank Kelly Hughes for suggestions and discussion and Fumio Oosawa and Sho Asakura for continuous support and encouragement.
This work was partially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (T.M. and K.N.).
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Asai, Y., T. Yakushi, I. Kawagishi, and M. Homma. 2003. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 327453-463. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi, T., Y. Maekawa, T. Yamada, I. Kawagishi, Y. Imae, and M. Homma. 1996. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J. Bacteriol. 1785024-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 7219-54. [DOI] [PubMed] [Google Scholar]
- 4.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60439-449. [DOI] [PubMed] [Google Scholar]
- 5.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 2421678-1681. [DOI] [PubMed] [Google Scholar]
- 6.Blair, D. F., D. Y. Kim, and H. C. Berg. 1991. Mutant MotB proteins in Escherichia coli. J. Bacteriol. 1734049-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block, S. M., and H. C. Berg. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309470-472. [DOI] [PubMed] [Google Scholar]
- 8.Braun, T. F., L. Q. Al-Mawsawi, S. Kojima, and D. F. Blair. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 4335-45. [DOI] [PubMed] [Google Scholar]
- 9.Braun, T. F., and D. F. Blair. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry 4013051-13059. [DOI] [PubMed] [Google Scholar]
- 10.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 213225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, P. N., M. A. Mathews, L. A. Joss, C. P. Hill, and D. F. Blair. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 1872890-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239276-278. [DOI] [PubMed] [Google Scholar]
- 13.Dean, G. D., R. M. Macnab, J. Stader, P. Matsumura, and C. Burks. 1984. Gene sequence and predicted amino acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12333-334. [DOI] [PubMed] [Google Scholar]
- 15.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 2351261-1270. [DOI] [PubMed] [Google Scholar]
- 16.Grizot, S., and S. K. Buchanan. 2004. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 511027-1037. [DOI] [PubMed] [Google Scholar]
- 17.Hosking, E. R., C. Vogt, E. P. Bakker, and M. D. Manson. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364921-937. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292195-202. [DOI] [PubMed] [Google Scholar]
- 19.Koebnik, R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 161269-1270. [DOI] [PubMed] [Google Scholar]
- 20.Kojima, S., and D. F. Blair. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 23393-134. [DOI] [PubMed] [Google Scholar]
- 21.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 4013041-13050. [DOI] [PubMed] [Google Scholar]
- 22.Kojima, S., and D. F. Blair. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 4326-34. [DOI] [PubMed] [Google Scholar]
- 23.Leake, M. C., J. H. Chandler, G. H. Wadhams, F. Bai, R. M. Berry, and J. P. Armitage. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443355-358. [DOI] [PubMed] [Google Scholar]
- 24.Lloyd, S. A., F. G. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400472-475. [DOI] [PubMed] [Google Scholar]
- 25.Lowder, B. J., M. D. Duyvesteyn, and D. F. Blair. 2005. FliG subunit arrangement in the flagellar rotor probed by targeted cross-linking. J. Bacteriol. 1875640-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 5777-100. [DOI] [PubMed] [Google Scholar]
- 27.Minamino, T., and K. Namba. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 75-17. [DOI] [PubMed] [Google Scholar]
- 28.Minamino, T., Y. Saijo-Hamano, Y. Furukawa, B. Gonzalez-Pedrajo, R. M. Macnab, and K. Namba. 2004. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J. Mol. Biol. 341491-502. [DOI] [PubMed] [Google Scholar]
- 29.Muramoto, K., and R. M. Macnab. 1998. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 291191-1202. [DOI] [PubMed] [Google Scholar]
- 30.Park, S.-Y., B. Lowder, A. M. Bilwes, D. F. Blair, and B. R. Crane. 2006. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. USA 10311886-11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons, L. M., F. Lin, and J. Orban. 2006. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 452122-2128. [DOI] [PubMed] [Google Scholar]
- 32.Paul, K., and D. F. Blair. 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J. Bacteriol. 1882502-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul, K., J. G. Harmon, and D. F. Blair. 2006. Mutational analysis of the flagellar rotor protein FliN: identification of surfaces important for flagellar assembly and switching. J. Bacteriol. 1885240-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid, S. W., M. C. Leake, J. H. Chandler, C. J. Lo, J. P. Armitage, and R. M. Berry. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA 1038066-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saijo-Hamano, Y., T. Minamino, R. M. Macnab, and K. Namba. 2004. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J. Mol. Biol. 343457-466. [DOI] [PubMed] [Google Scholar]
- 36.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 2755718-5722. [DOI] [PubMed] [Google Scholar]
- 37.Sato, K., and M. Homma. 2000. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. J. Biol. Chem. 27520223-20228. [DOI] [PubMed] [Google Scholar]
- 38.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 39.Sowa, Y., A. D. Rowe, M. C. Leake, T. Yakushi, M. Homma, A. Ishijima, and R. M. Berry. 2005. Direct observation of steps in rotation of the bacterial flagellar motor. Nature 437916-919. [DOI] [PubMed] [Google Scholar]
- 40.Stader, J., P. Matsumura, D. Vacante, G. E. Dean, and R. M. Macnab. 1986. Nucleotide sequence of the Escherichia coli MotB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 166244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolz, B., and H. C. Berg. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 1737033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 1787069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yakushi, T., N. Hattori, and M. Homma. 2005. Deletion analysis of the carboxyl-terminal region of the PomB component of the Vibrio alginolyticus polar flagellar motor. J. Bacteriol. 187778-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi, S., S. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 1681172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130255-265. [DOI] [PubMed] [Google Scholar]
- 47.Yorimitsu, T., and M. Homma. 2001. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 150582-93. [DOI] [PubMed] [Google Scholar]
- 48.Yorimitsu, T., M. Kojima, T. Yakushi, and M. Homma. 2004. Multimeric structure of the PomA/PomB channel complex in the Na+-driven flagellar motor of Vibrio alginolyticus. J. Biochem. (Tokyo) 13543-51. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., R. T. Fazzio, and D. F. Blair. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251237-242. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 956436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 1802729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]