Abstract
The putative global posttranscriptional regulator csrA was mutated in Campylobacter jejuni 81-176. The csrA mutant was attenuated in surviving oxidative stress. CsrA also contributed to biofilm formation and adherence to and invasion of INT407 intestinal epithelial cells, suggesting a regulatory role for CsrA in C. jejuni pathogenesis.
Diarrheal diseases represent an immense burden to both the developing and the industrial world, and the gram-negative pathogen Campylobacter jejuni is recognized around the world as a leading bacterial cause of gastroenteritis (3, 13, 16). Although C. jejuni requires very specific growth conditions in the laboratory, it persists in the environment. As it passes from host (commonly avian species) to human, C. jejuni must survive a great range of hostile environmental stresses, including limited carbon sources, suboptimal growth temperatures, and exposure to atmospheric oxygen. During infection, C. jejuni must withstand changes in pH and the host inflammatory response. In order to survive these stresses, C. jejuni must be able to sense these changes and respond accordingly. However, relatively little is known about the molecular mechanisms of Campylobacter pathogenesis and even less is known about how its virulence properties are regulated. While C. jejuni possesses several predicted global regulatory systems, including regulators of flagellar assembly and function (28, 67), iron homeostasis (58), heat shock (33), cold shock (45; W. A. Agee and S. A. Thompson, unpublished data), and the stringent response (19), its complement of regulators is dramatically less than that of enteric pathogens such as Salmonella enterica. Furthermore, C. jejuni has only three sigma factors (σ70 [rpoD], σ54 [rpoN], and σ28 [fliA]), seven histidine kinases, and 10 response regulators (44, 45). The small number of sigma factors and other global regulators in C. jejuni suggests that there may be other uncharacterized mechanisms of gene regulation.
C. jejuni genome sequences (18, 45) revealed orthologs of the Escherichia coli global posttranscriptional regulator csrA (carbon starvation regulator). In E. coli, CsrA was identified as a posttranscriptional regulator of translation (49, 50) responsible for repression or activation of many important processes. CsrA is a homodimeric RNA-binding protein that typically binds the 5′ untranslated regions of target mRNAs at one or more sites that are often adjacent to or overlapping the ribosome binding site, thus inhibiting ribosome access to the ribosome binding site and inhibiting translation initiation, which can either increase or decrease mRNA half-life (5, 7, 15, 39, 40, 48, 61).
In E. coli, CsrA is involved in regulating stationary-phase metabolism, represses glycogen biosynthesis, gluconeogenesis, peptide transport, and biofilm formation (2, 15, 27, 37, 51, 52, 61), and activates glycolysis, acetate metabolism, and motility (52, 63, 64). Analysis of bacterial genomes has revealed widespread distribution of csrA throughout the eubacteria (65). Subsequently, the role of CsrA in the life cycles of several pathogenic bacteria has been studied, revealing that CsrA not only regulates stationary-phase metabolism but also is an important regulator of virulence determinants, including host cell invasion, quorum sensing, biofilm formation, iron acquisition, type III secretion systems, and outer membrane protein expression (4, 11, 12, 17, 25, 26, 34, 37, 38, 42, 43, 46, 47, 66). In the gastric pathogen Helicobacter pylori, a close relative of C. jejuni (21), CsrA is reported to play a role in the regulation of several virulence phenotypes, including motility, oxidative stress resistance, and mouse colonization (8).
Considering the limited contingent of regulatory effectors found in C. jejuni genomes, we suspected that CsrA might play a vital role in the regulation of stress responses and virulence determinants in this enteric pathogen. In this study, we sought to examine the role of CsrA in C. jejuni pathogenesis. We therefore constructed a C. jejuni 81-176 csrA mutant and complemented mutant strains for use in studies of survival and virulence-related phenotypes. We report that mutation of csrA reveals a potential role for CsrA in the regulation of C. jejuni genes required for survival of oxidative stress. Furthermore, CsrA plays a role in the activation of biofilm formation, motility, and adherence to host cells in vitro; however, it contributes to the repression of invasion of human cells.
Mutation of csrA in C. jejuni 81-176.
A nonpolar mutation in csrA was constructed by inverse-PCR mutagenesis (68). Briefly, by use of primers JAF44 and JAF45, Cj1103 (csrA) including 500 bp upstream and downstream was amplified using PCR and cloned into pCRII-TOPO (Invitrogen). The new construct, pJF06, was then subjected to inverse PCR using primers JAF50A and JAF51, digested with NheI, and self-ligated to yield pJF07. pJF07 was digested with NheI and XmaI and ligated with the chloramphenicol acetyltransferase (cat) cassette amplified from pRY111 (69) by use of primers JAF52 and JAF53 and digested with the same enzymes to generate the plasmid pJF09. This plasmid contained a deletion of 75% of the csrA gene (replaced with cat) while maintaining the translation initiation signals of the downstream Cj1104 gene to avoid polarity. This construct was then introduced into C. jejuni 81-176 by electroporation (62), and a chloramphenicol-resistant (20 μg/ml) csrA mutant was verified by PCR and DNA sequencing (data not shown).
Complementation of the csrA mutant in trans.
Complementation of the csrA mutant was accomplished by introducing the csrA gene under the control of its native promoter on the Campylobacter shuttle vector pRY107 (69). Briefly, csrA was amplified with primers JAF60 and JAF43 and cloned into pCRII-TOPO, producing pJF10A. Next, the csrA promoter (upstream of Cj1097) was amplified with primers JAF61 and JAF62, digested with XmaI and NdeI, and cloned upstream of csrA in pJF10A to create pJF10B. The csrA promoter cassette was then digested with EcoRI and subcloned into pRY107, giving the csrA complementation vector pJF11. pJF11 was then introduced into the csrA mutant by triparental mating (36). Transconjugants were recovered on chloramphenicol (15 μg/ml) and kanamycin (50 μg/ml), and the presence of pJF11 was confirmed by plasmid midi-prep (Qiagen) (data not shown).
Mutation of csrA decreases swarming ability.
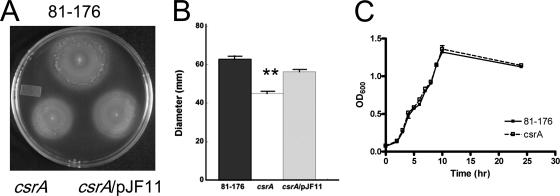
The swarming ability of the csrA mutant was determined on Mueller-Hinton (MH) media containing 0.4% agar (22) and confirmed via light microscopy of wet mounts (data not shown). After inoculation, the strains were incubated at 37°C for 24 h (Fig. 1A) and 48 h (Fig. 1B). The swarming ability of the mutant was >30% less than that of the parent strain after 24 h (P = 0.009) and 48 h (P = 0.0007), despite highly similar growth characteristics in MH broth (Fig. 1C). This was consistent with reported observations for E. coli and H. pylori (8, 64) and suggests that C. jejuni CsrA contributes to the regulation of motility or chemotaxis, as either can affect swarming ability.
FIG. 1.
CsrA is required for full motility. Swarming ability was assessed on MH agar containing 0.4% agar. Strains were inoculated into MH motility agar and incubated for 24 h (A) and 48 h (B) at 37°C under microaerobic conditions. (C) Growth of the 81-176 and 81-176 csrA strains was observed in MH broth and measured by determining the OD600. The assay was carried out in triplicate, and one representative of three experiments is shown (**, P ≤ 0.005) with error bars.
CsrA is required for resistance to oxidative stress.
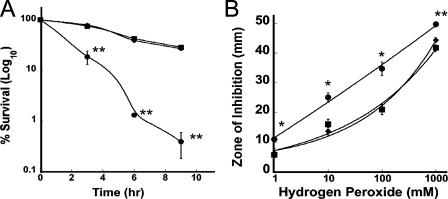
Resistance of the 81-176, 81-176 csrA, and 81-176 csrA/pJF11 strains to oxidative stress was determined by assessing killing by atmospheric oxygen (19) and hydrogen peroxide (60). Aerotolerance was determined by transferring bacteria grown in MH broth to early log phase (optical density at 600 nm [OD600] of ∼0.1) from a microaerobic environment to atmospheric and microaerobic growth conditions and incubating the bacteria for 9 h at 37°C. At 0, 3, 6, and 9 h, viable counts were measured by serial dilution and plating on MH plates. This experiment (Fig. 2A) showed that the csrA mutant was highly sensitive to atmospheric oxygen, resulting in greater than 99% loss of viability by 9 h (P = 0.0005). The strains grown under microaerobic conditions remained viable and grew to stationary phase (data not shown), indicating that the loss of viability under atmospheric conditions was specific to atmospheric oxygen exposure. For hydrogen peroxide resistance, cells were grown on blood agar overnight at 37°C, harvested in phosphate-buffered saline, and diluted to an OD600 of ∼1.0. A 100-μl portion of each strain was spread on MH agar, onto which filter discs (6 mm) inoculated with 10 μl of 1 mM, 10 mM, 100 mM, or 1 M hydrogen peroxide were placed and then incubated at 37°C under microaerobic conditions for 48 h. These studies (Fig. 2B) revealed greater sensitivity of the csrA mutant to all doses tested (P ≤ 0.01). Taken together these data suggest that, as in H. pylori, CsrA contributes to the regulation of oxidative stress responses in C. jejuni.
FIG. 2.
Deletion of CsrA results in reduced resistance to sources of oxidative stress. The 81-176 (▪), 81-176 csrA (•), and 81-176 csrA/pJF11 (⧫) strains were subjected to oxidative stress by exposure to atmospheric oxygen, whereby the strains were inoculated in flasks at an OD600 of ∼0.1 and a 6:1 surface-to-volume ratio and then incubated at 37°C and 100 rpm in an air incubator (A), and various concentrations of hydrogen peroxide in filter discs on MH agar plates (B). One representative, in triplicate, of three experiments is shown (*, P ≤ 0.05; **, P ≤ 0.005) with error bars.
CsrA is an activator of biofilm formation.
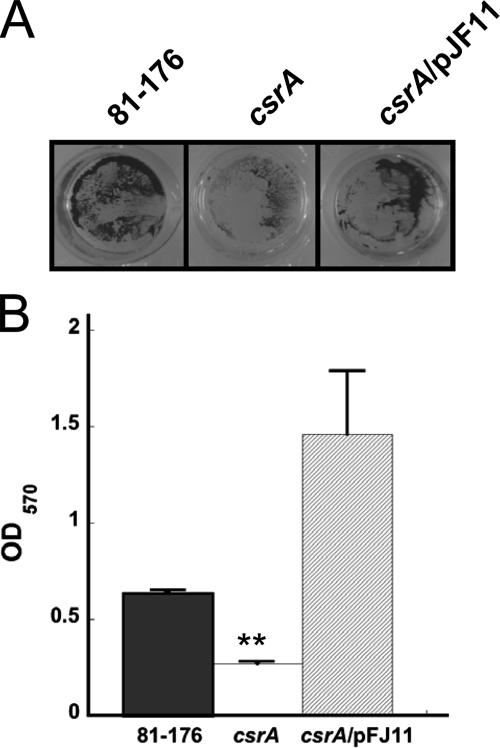
By use of previously described methods (14), biofilms were quantitated via crystal violet (CV) staining of static biofilm formation in 24-well, flat-bottomed polystyrene tissue culture dishes at 48 h. Briefly, strains were inoculated in MH broth at an OD600 of 0.05 and incubated statically at 37°C for 48 h. Biofilms were visualized by staining with CV, washed with distilled H2O, and photographed, and CV binding was quantitated by determining the OD570 after solubilization in 80% dimethyl sulfoxide for 24 h (Fig. 3). The csrA mutant formed a very sparse biofilm on the bottoms and sides of the wells (Fig. 3A). Conversely, both the wild type and the complement formed dense biofilms; however, a great deal of the matrix formed by the complemented mutant was present on the sides of the wells and is not represented in the aspect shown. Quantification of CV staining (Fig. 3B) revealed that the csrA mutant formed nearly 50% less biofilm than 81-176 (P = 0.0001); however, the complemented mutant formed twice as much biofilm as the wild type. It has been demonstrated that flagellar function and responses to both general and oxidative stress are critical to biofilm formation (24, 30, 31, 57, 59). These results suggest that CsrA is an activator of biofilm formation, possibly via regulation of motility and oxidative stress responses in C. jejuni. This conclusion is noteworthy considering that CsrA represses biofilm formation in several gammaproteobacteria (1, 2, 27, 54, 61). Biofilm formation in C. jejuni is poorly understood but is certainly complex and requires flagellar function (30). Therefore, reduced biofilm formation by the C. jejuni CsrA mutant is consistent with the observation of reduced motility (Fig. 1) and also suggests that CsrA-mediated control of biofilm formation may be inherently different in C. jejuni and E. coli.
FIG. 3.
CV staining of C. jejuni biofilms. CV-stained biofilms were solubilized in 80% dimethyl sulfoxide (A) and quantitated by determining the OD570 (B). One representative, in triplicate, of three experiments is shown (**, P ≤ 0.005) with error bars.
Adherence and invasion of intestinal epithelial cells.
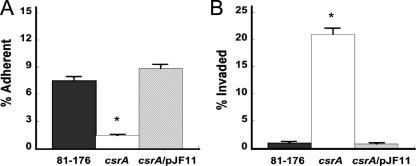
The role of CsrA in adherence and invasion of host cells in vitro was determined as previously described (9, 41, 62). The csrA mutant exhibited a 5.4-fold decrease in the ability to adhere to INT407 cells (Fig. 4A) (P = 0.002). This attenuation of adherence was contrasted by a 20-fold increase in invasion by adherent C. jejuni csrA mutant cells (Fig. 4B) (P = 0.01) despite reduced motility, a factor known to influence invasion (20, 23, 29, 70). There was no difference in susceptibility to gentamicin among the strains. This is the first report to implicate CsrA in the regulation of host cell adherence. Previous studies have reported that CsrA functions in both the activation and the repression of invasion (4, 17, 37). Our data suggest that in C. jejuni the role of CsrA in epithelial cell invasion is primarily carried out via repression of invasion-specific genes. This conclusion introduces a paradox because both motility and adherence are important for host cell invasion in C. jejuni (20, 23, 29, 62, 70); however, the csrA mutant displays defects in both. However, while both motility and adherence are certainly prerequisites for invasion, the adherence and invasion processes involve different proteins. For example, molecules that are involved uniquely in the invasion step but not in adherence include the Campylobacter invasion antigens, gamma-glutamyl transpeptidase, and the polysaccharide capsule (6, 9, 32). CsrA may therefore directly or indirectly regulate these or other invasion-specific Campylobacter proteins, and changes in the expression of these proteins may override any effect of the decrease in motility and result in the observed increase in invasion.
FIG. 4.
Adherence and invasion of INT407 cells. The capacities of the 81-176, 81-176 csrA, and 81-176 csrA/pJF11 strains to adhere to and invade INT407 cells at a multiplicity of infection of 15 were examined in vitro. (A) Adherence is expressed as the percentage of bacteria which had either adhered to or invaded cultured intestinal epithelial cells after a 3-h incubation, compared to the inoculum. (B) Invasiveness is expressed as the percentage of intracellular bacteria surviving gentamicin treatment of the INT407 cells after an additional 2-h incubation, compared to the number of adherent bacteria (to account for differences in adherence among strains). One representative, in triplicate, of three experiments is shown (*, P ≤ 0.05) with error bars.
Conclusions.
Posttranscriptional regulation in C. jejuni has not been studied previously, and many questions remain to be considered in future studies to address how CsrA works in Campylobacter and other epsilonproteobacteria. Presently, it is not known how CsrA is regulated in C. jejuni. In E. coli and many other bacteria, CsrA has been shown to be regulated by the small noncoding RNAs csrB and csrC (5), which have not been identified in Campylobacter (35). Furthermore, regulation of E. coli csrBC is directed by the BarA/UvrY two-component regulatory system (17, 53-56), which does not appear to have an ortholog in C. jejuni (45). These data, therefore, represent an important first step in elucidating the role of CsrA in C. jejuni physiology and pathogenesis. In summary, we have constructed a C. jejuni mutant lacking the predicted posttranscriptional regulator CsrA. The csrA mutant exhibits changes in several virulence-related properties, including oxidative stress resistance, motility, adherence, and invasion. These pleiotropic effects suggest that CsrA is an important regulator involved in C. jejuni pathogenesis.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Resistancea | Source or reference |
|---|---|---|---|
| Strains | |||
| E. coli JM109 | Cloning host | Promega | |
| C. jejuni 81-176 | Wild type | 10 | |
| Plasmids | |||
| pCRII-TOPO | Cloning vector | Amp, Kan | Invitrogen |
| pRY107 | C. jejuni shuttle vector | Kan | 69 |
| pRY111 | C. jejuni shuttle vector | Cm | 69 |
| pJF06 | 1.2-kb csrA locus in pCRII-TOPO | Amp, Kan | This study |
| pJF07 | Self-ligated inverse-PCR product of pJF06 | Amp, Kan | This study |
| pJF09 | pJF07::csrAΔcat | Amp, Kan, Cm | This study |
| pJF10A | csrA in pCRII-TOPO | Kan | This study |
| pJF10B | pJF10A and pcsrA | Kan | This study |
| pJF11 | csrA and promoter in pRY107 | Kan | This study |
Amp, ampicillin; Kan, kanamycin; Cm, chloramphenicol.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ → 3′)a |
|---|---|
| JAF43 | TCA TTT GAT TAG TTT TTT GC |
| JAF44 | ATG CAA GGA ATT ATC TCC TA |
| JAF45 | GGT ATG TCA TCT TCA AAT TC |
| JAF50A | CTC TGC TAG CAC CCG GGT GTT GTT CAG AAT GAT ATT AAA C |
| JAF51 | AGA GGC TAG CTT AAC ATT TTT CAA CCT TAT T |
| JAF52 | CTC TGC TAG CGG AGG ATA AAT GAT GCA ATT |
| JAF53 | AGA GCC CGG GTT ATT TAT TCA GCA AGT CTT |
| JAF60 | CTA CCC GGG ATT CAT ATG TTA ATA TTA TCA |
| JAF61 | GAT CCC GGG TAA TCA GCT TTA CTA AGT TTG TGA TTT GAC |
| JAF62 | GCT CAT ATG AAA AAC CTT ATT AAA TAT TTT TTA TAT CAA AAG |
Underlined nucleotides indicate restriction sites introduced for cloning purposes.
Acknowledgments
This study was supported by National Institutes of Health grants AI055715 and AI058284 to S.A.T.
We thank Qijing Zhang (Iowa State University) for plasmid pRK212.1 and the Thompson laboratory for critical reading of the manuscript.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Agladze, K., D. Jackson, and T. Romeo. 2003. Periodicity of cell attachment patterns during Escherichia coli biofilm development. J. Bacteriol. 1855632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agladze, K., X. Wang, and T. Romeo. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 1878237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 321201-1206. [DOI] [PubMed] [Google Scholar]
- 4.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 686790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babitzke, P., and T. Romeo. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10156-163. [DOI] [PubMed] [Google Scholar]
- 6.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40769-777. [DOI] [PubMed] [Google Scholar]
- 7.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 441599-1610. [DOI] [PubMed] [Google Scholar]
- 8.Barnard, F. M., M. F. Loughlin, H. P. Fainberg, M. P. Messenger, D. W. Ussery, P. Williams, and P. J. Jenks. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 5115-32. [DOI] [PubMed] [Google Scholar]
- 9.Barnes, I. H., M. C. Bagnall, D. D. Browning, S. A. Thompson, G. Manning, and D. G. Newell. 2007. Gamma-glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb. Pathog. 43198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157472-479. [DOI] [PubMed] [Google Scholar]
- 11.Burrowes, E., A. Abbas, A. O'Neill, C. Adams, and F. O'Gara. 2005. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 1567-16. [DOI] [PubMed] [Google Scholar]
- 12.Burrowes, E., C. Baysse, C. Adams, and F. O'Gara. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152405-418. [DOI] [PubMed] [Google Scholar]
- 13.Butzler, J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10868-876. [DOI] [PubMed] [Google Scholar]
- 14.Candon, H. L., B. J. Allan, C. D. Fraley, and E. C. Gaynor. 2007. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 1898099-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubey, A. K., C. S. Baker, K. Suzuki, A. D. Jones, P. Pandit, T. Romeo, and P. Babitzke. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 1854450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields, P. I., and D. L. Swerdlow. 1999. Campylobacter jejuni. Clin. Lab. Med. 19489-504. [PubMed] [Google Scholar]
- 17.Fortune, D. R., M. Suyemoto, and C. Altier. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 74331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 568-27. [DOI] [PubMed] [Google Scholar]
- 20.Golden, N. J., and D. W. Acheson. 2002. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 701761-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin, C. S., and J. A. Armstrong. 1990. Microbiological aspects of Helicobacter pylori (Campylobacter pylori). Eur. J. Clin. Microbiol. Infect. Dis. 91-13. [DOI] [PubMed] [Google Scholar]
- 22.Goossens, H., L. Vlaes, I. Galand, C. Van den Borre, and J. P. Butzler. 1989. Semisolid blood-free selective-motility medium for the isolation of campylobacters from stool specimens. J. Clin. Microbiol. 271077-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, C. C., M. E. Konkel, W. Cieplak, Jr., and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 611764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19417-428. [DOI] [PubMed] [Google Scholar]
- 25.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Camara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 1862936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins, D. A., M. E. Pomianek, C. M. Kraml, R. K. Taylor, M. F. Semmelhack, and B. L. Bassler. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450883-886. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 1832937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakuda, T., and V. J. DiRita. 2006. Cj1496c encodes a Campylobacter jejuni glycoprotein that influences invasion of human epithelial cells and colonization of the chick gastrointestinal tract. Infect. Immun. 744715-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalmokoff, M., P. Lanthier, T. L. Tremblay, M. Foss, P. C. Lau, G. Sanders, J. Austin, J. Kelly, and C. M. Szymanski. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 1884312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirov, S. M., M. Castrisios, and J. G. Shaw. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 721939-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konkel, M. E., B. J. Kim, V. Rivera-Amill, and S. G. Garvis. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32691-701. [DOI] [PubMed] [Google Scholar]
- 33.Konkel, M. E., R. T. Marconi, D. J. Mead, and W. Cieplak, Jr. 1994. Cloning and expression of the hup encoding a histone-like protein of Campylobacter jejuni. Gene 14683-86. [DOI] [PubMed] [Google Scholar]
- 34.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6186-190. [DOI] [PubMed] [Google Scholar]
- 35.Kulkarni, P. R., X. Cui, J. W. Williams, A. M. Stevens, and R. V. Kulkarni. 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 343361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labigne-Roussel, A., J. Harel, and L. Tompkins. 1987. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 1695320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawhon, S. D., J. G. Frye, M. Suyemoto, S. Porwollik, M. McClelland, and C. Altier. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 481633-1645. [DOI] [PubMed] [Google Scholar]
- 38.Lenz, D. H., and B. L. Bassler. 2007. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 63859-871. [DOI] [PubMed] [Google Scholar]
- 39.Liu, M. Y., H. Yang, and T. Romeo. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 1772663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 4093-113. [DOI] [PubMed] [Google Scholar]
- 41.Monteville, M. R., J. E. Yoon, and M. E. Konkel. 2003. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 149153-165. [DOI] [PubMed] [Google Scholar]
- 42.Mulcahy, H., J. O'Callaghan, E. P. O'Grady, C. Adams, and F. O'Gara. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 743012-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Grady, E. P., H. Mulcahy, J. O'Callaghan, C. Adams, and F. O'Gara. 2006. Pseudomonas aeruginosa infection of airway epithelial cells modulates expression of Kruppel-like factors 2 and 6 via RsmA-mediated regulation of type III exoenzymes S and Y. Infect. Immun. 745893-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, S. 2000. Environmental regulatory genes, p. 423-440. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 45.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 46.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 1836676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raffatellu, M., R. P. Wilson, D. Chessa, H. Andrews-Polymenis, Q. T. Tran, S. Lawhon, S. Khare, L. G. Adams, and A. J. Bäumler. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73146-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 291321-1330. [DOI] [PubMed] [Google Scholar]
- 49.Romeo, T. 1996. Post-transcriptional regulation of bacterial carbohydrate metabolism: evidence that the gene product CsrA is a global mRNA decay factor. Res. Microbiol. 147505-512. [DOI] [PubMed] [Google Scholar]
- 50.Romeo, T., and M. Gong. 1993. Genetic and physical mapping of the regulatory gene csrA on the Escherichia coli K-12 chromosome. J. Bacteriol. 1755740-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1754744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabnis, N. A., H. Yang, and T. Romeo. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 27029096-29104. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 1845130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teplitski, M., A. Al-Agely, and B. M. Ahmer. 2006. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology 1523411-3424. [DOI] [PubMed] [Google Scholar]
- 55.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2006. Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int. J. Med. Microbiol. 296449-466. [DOI] [PubMed] [Google Scholar]
- 56.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 1857257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26173-186. [DOI] [PubMed] [Google Scholar]
- 58.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 1805291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vatanyoopaisarn, S., A. Nazli, C. E. Dodd, C. E. Rees, and W. M. Waites. 2000. Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl. Environ. Microbiol. 66860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wai, S. N., K. Nakayama, K. Umene, T. Moriya, and K. Amako. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 201127-1134. [DOI] [PubMed] [Google Scholar]
- 61.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 561648-1663. [DOI] [PubMed] [Google Scholar]
- 62.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 102055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei, B., S. Shin, D. LaPorte, A. J. Wolfe, and T. Romeo. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 1821632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40245-256. [DOI] [PubMed] [Google Scholar]
- 65.White, D., M. E. Hart, and T. Romeo. 1996. Phylogenetic distribution of the global regulatory gene csrA among eubacteria. Gene 182221-223. [DOI] [PubMed] [Google Scholar]
- 66.Wood, M. W., M. A. Jones, P. R. Watson, A. M. Siber, B. A. McCormick, S. Hedges, R. Rosqvist, T. S. Wallis, and E. E. Galyov. 2000. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell. Microbiol. 2293-303. [DOI] [PubMed] [Google Scholar]
- 67.Wosten, M. M., J. A. Wagenaar, and J. P. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 27916214-16222. [DOI] [PubMed] [Google Scholar]
- 68.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16994-996. [PubMed] [Google Scholar]
- 69.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130127-130. [DOI] [PubMed] [Google Scholar]
- 70.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and nonmotile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14883-893. [DOI] [PubMed] [Google Scholar]