Abstract
Lactobacillus johnsonii strains NCC533 and ATCC 33200 (the type strain of this species) differed significantly in gut residence time (12 versus 5 days) after oral feeding to mice. Genes affecting the long gut residence time of the probiotic strain NCC533 were targeted for analysis. We hypothesized that genes specific for this strain, which are expressed during passage of the bacterium through the gut, affect the phenotype. When the DNA of the type strain was hybridized against a microarray of the sequenced NCC533 strain, we identified 233 genes that were specific for the long-gut-persistence isolate. Whole-genome transcription analysis of the NCC533 strain using the microarray format identified 174 genes that were strongly and consistently expressed in the jejunum of mice monocolonized with this strain. Fusion of the two microarray data sets identified three gene loci that were both expressed in vivo and specific to the long-gut-persistence isolate. The identified genes included LJ1027 and LJ1028, two glycosyltransferase genes in the exopolysaccharide synthesis operon; LJ1654 to LJ1656, encoding a sugar phosphotransferase system (PTS) transporter annotated as mannose PTS; and LJ1680, whose product shares 30% amino acid identity with immunoglobulin A proteases from pathogenic bacteria. Knockout mutants were tested in vivo. The experiments revealed that deletion of LJ1654 to LJ1656 and LJ1680 decreased the gut residence time, while a mutant with a deleted exopolysaccharide biosynthesis cluster had a slightly increased residence time.
Gut bacteria are of fundamental interest for microbiologists and nutritionists since pathogens exploit their hosts for nutrients, commensals are nutritionally neutral for their hosts, and symbionts contribute to the nutrition of their hosts by digesting dietary fiber (54, 55). An understanding of the microbe-host interactions in the gut is thus not only of fundamental interest but also of medical interest. Some gut commensals might contribute to the development of obesity (45), while others protect the host against infection with pathogens (1, 4). In fact, gut commensals like Escherichia coli (34), lactobacilli (47), and bifidobacteria (36) have been administered orally for treatment of diarrheal diseases and inflammatory conditions. Lactobacilli, which represent an important part of the natural gut microbiome in both humans and animals, including laboratory mice (42), have been intensively explored as probiotics (i.e., health-promoting bacteria) (19). However, the genetic basis of probiotic effects has been only poorly defined (38). Obviously, the complexity of the gut microbiome makes the genetic approach to defining microbe-microbe and microbe-host interactions challenging.
In the present study we tried to identify genes in a probiotic Lactobacillus johnsonii strain (13, 15, 33) which affect a physiologically relevant in vivo trait, gut persistence. It is commonly assumed that a successful probiotic bacterium must achieve and maintain a sizable intestinal cell titer after oral application to have an effect (11, 44). A strain persisting in the gut must resist stomach acidity, proteases, bile acids, and lipases. In addition, it must develop some basic metabolic activity in the gut and resist the selection pressure of immunoglobulin A (IgA), which is abundantly secreted into the intestine.
While this property is a necessary but not sufficient requirement for probiotic activity, strains that transit only passively through the gut or do not cross a critical threshold in the gut cell population (about 106 bacteria/g feces) are not selected in industrial screening programs for probiotics. In mouse feeding experiments the probiotic L. johnsonii strain NCC533 selected in our laboratory (33) showed an intestinal persistence with elevated fecal cell titers that was about 10 days longer than that of the L. johnsonii type strain, strain ATCC 33200. It should be noted that persistence of bacteria in the gut has two meanings (42). Autochthonous species have a long-term association with particular host species, forming a stable population of a characteristic size in a particular region of the gut, and have a demonstrable ecological function. In contrast, allochthonous species are commonly introduced into the gut ecosystem because they are ubiquitous in nature and are part of the feed or food or are deliberately introduced as probiotics. Experiments with our L. johnsonii strains and previous experiments with other probiotic lactobacilli (28) have addressed the second situation. Thus, the distinction between the “long-gut-persistence” phenotype and the “short-gut-persistence” phenotype refers only to a quantitative difference and not to a qualitative difference between strains (39).
In our laboratory the “long-gut-persistence” strain NCC533 was sequenced (33), and the genetic diversity of L. johnsonii strains was explored by genotyping with microarrays (3). This analysis revealed that in L. johnsonii the differences between strains involved up to 15% of the gene content, consistent with previous analyses of Lactobacillus plantarum (25). Some gene differences are simply “genetic noise” introduced by selfish genetic elements (49), while others truly reflect adaptations to a specific ecological niche.
How can physiologically relevant genes be filtered out of the strain-specific gene pool? Dutch scientists determined the gene conservation pattern of many L. plantarum strains by using genotyping microarrays (25), and they correlated this conservation pattern with an in vitro binding phenotype (mannose binding on yeast cells) and associated a large cell wall-anchored surface protein with the mannose-binding phenotype (31). However, in a mutant of L. plantarum with a sortase mutation, which should have interfered with the appropriate surface exposure of such an adhesin, the mutation did not affect the gut persistence phenotype of L. plantarum (31). In vivo expression technology (IVET) is an alternative approach to elucidate genes involved in gut persistence. This technique identifies bacterial genes that are specifically transcribed during gut passage in mice. With this approach three such genes were identified in Lactobacillus reuteri (52), but only one gene, the gene encoding the methionine sulfoxide reductase, had a small effect on the gut colonization phenotype when knockout (KO) mutants were used (51). In L. plantarum the IVET approach identified 72 genes which are specifically transcribed in the mouse gut (5). KO mutants with mutations in nine of these genes were constructed, and their gut passage in mice was tested. For three KO mutants the fecal excretion was 100- to 1,000-fold less than that of the isogenic wild-type strain. The identified genes were genes encoding a copper-transporting ATPase, a component of the cellobiose phosphotransferase transport system (PTS) (IIC transport component), and a hypothetical extracellular protein (7).
Here we hypothesized that only strain-specific genes which are expressed in the gut are likely to affect the investigated phenotype. Below we propose a method to identify relevant genes by using two phenotypically distinct strains and a combination of genotyping and expression microarray analyses. We tested the method for the “long-gut-persistence” phenotype displayed by the probiotic strain NCC533. KO mutants of NCC533 revealed that two of the three identified genetic loci were indeed associated with reduced gut residence times in mice. We suggest that this approach represents a promising tool to elucidate the genetic basis of complex bacterial phenotypes in mammalian hosts.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. johnsonii strains ATCC 33200T and NCC533 were grown in MRS with 2% glucose and 0.05% (wt/vol) cysteine at 37°C under anaerobic conditions. Lactococcus lactis strain MG1363 was grown at 30°C in M17-glucose. Escherichia coli K-12 strain K803 was grown in brain heart infusion or LB medium at 37°C with shaking.
Cells were grown in a Sixfors fermentor system composed of four individual 500-ml vessels (Infors, Bottmingen, Switzerland). Anaerobic conditions were obtained by flushing the medium with CO2 prior to inoculation. All fermentations were carried out at 37°C with constant agitation (150 rpm). The starting optical density at 600 nm was 0.05.
Antibiotics were used at the following concentrations: erythromycin, 10 μg/ml for Lactobacillus and Lactococcus and 150 μg/ml for E. coli; and chloramphenicol, 10 μg/ml for Lactobacillus and Lactococcus and 25 μg/ml for E. coli. MRS is relatively specific for lactobacilli. Mouse feces yield high titers of endogenous lactobacilli on MRS. However, in the presence of the antibiotics specified above no colonies were detected in the feces of normal mice.
The stability of the plasmids was tested by using 20 serial in vitro passages in the absence of antibiotics. Less than 1% antibiotic-sensitive strains were observed. All strains used in this study are described in Table S1 in the supplemental material.
Electron microscopy.
Cells were centrifuged and fixed by immersion in a solution of 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4). The liquid samples were encapsulated in agar gel tubes and postfixed with 2% osmium tetroxide in the same buffer; both solutions contained 0.04% ruthenium red. After dehydration in a graded ethanol series (70, 80, 90, 96, and 100% ethanol), the samples were embedded in Spurr resin. After polymerization of the resin (70°C, 48 h), ultrathin sections were cut with a Reichert OMU2 ultramicrotome and then stained with aqueous uranyl acetate and lead citrate. The sections were examined by transmission electron microscopy (Philips CM12) at 80 kV.
Gene inactivation.
General molecular cloning techniques, restriction enzyme analysis, and transformation of E. coli by CaCl2-induced competence were performed using standard procedures. Plasmid DNA from E. coli and Lactobacillus was isolated with a QIAprep spin miniprep kit (Qiagen). Recombinant DNA molecules were introduced into L. lactis and Lactobacillus by electrotransformation. Gene inactivation was achieved by allele exchange replacement using the pDP600 plasmid (Cmr Emr). Flanking regions of the target genes were amplified and inserted into the plasmid by directional cloning in E. coli (see Fig. S1 in the supplemental material). All constructs were checked by PCR using primers flanking the target region (see Table S2 in the supplemental material).
All primers used for construction of the gene inactivation mutants are listed in Table S2 in the supplemental material. For this work we engineered a chloramphenicol resistance gene for use in Lactobacillus species by cloning a Lactobacillus promoter before the pNZ124 (29) chloramphenicol resistance gene and adding an L. lactis stem-loop terminator for efficient, single-copy selection in Lactobacillus. The 230-bp amplicon containing the promoter of the Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 lacS gene (18) was amplified using Pwo polymerase (Roche Diagnostics, Rotkreuz, Switzerland) and primers A-5′ and A-3′ introducing RcaI and EcoRI restriction sites, as shown in Fig. S2 in the supplemental material. The PCR was performed with an Applied Biosystems 9700 thermocycler (Applied Biosystems, Rotkreuz, Switzerland) using the following amplification protocol: heating at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, and 68°C for 2 min and finally incubation at 68°C for 7 min before holding at 4°C. The chloramphenicol resistance gene from plasmid pNZ124 was amplified with Pwo and primers B-5′ and B-3′ introducing RcaI and ClaI restriction sites. The two amplicons were digested with the corresponding enzymes and ligated together in the cloning vector pBluescript pBS SK+ (Stratagene) digested with EcoRI plus ClaI, the ligated construct was used to transform E. coli XL1-Blue, and chloramphenicol-resistant clones were selected. The stem-loop terminator from the L. lactis strain NCDO 2054 lactose-galactose operon (48) was amplified with Pwo using primers C-5′ and C-3′ introducing AsuII and SacI restriction sites. The 170-bp amplicon was digested with AsuII plus SacI and ligated to the EcoRI-ClaI fragment containing the chloramphenicol resistance gene plus the promoter in the cloning vector pK19 (32) digested with EcoRI plus SacI, resulting in plasmid pDP352. The pDP352 chloramphenicol resistance gene was amplified with Pwo using primer D-5′ introducing SacI and BglII sites and primer D-3′ introducing a SacI site. The amplicon was digested with SacI and ligated into pBluescript SK+ digested with SacI. Colonies were screened to identify a clone releasing the chloramphenicol resistance gene as a BglII-KpnI fragment, which was ligated to the pG+host9 (23) replicon after digestion with BamHI and KpnI to obtain plasmid pDP600, a chloramphenicol-resistant version of pG+host9 containing a complete pBluescript array of unique restriction sites. A DNA fragment containing the erythromycin resistance gene was amplified from plasmid pVA838 (22) using primers E-5′ and E-3′ introducing PstI and EcoRI restriction sites, respectively. The amplicon was digested with PstI plus EcoRI and ligated into pDP600 digested with PstI plus EcoRI to produce pDP600-Ery.
The targeted region 3′ and 5′ homologous sequences were amplified with Pwo from NCC533 chromosomal DNA using specific PCR primers (see Table S2 in the supplemental material). The amplicon was digested and ligated into the plasmid described above digested with the same enzymes to obtain plasmid pDP735. Plasmid pDP735 isolated from MG1363 was used to transform NCC533 and was selected on MRS plates supplemented with erythromycin at 32°C (permissive temperature for plasmid replication). A culture from a single colony in MRS broth supplemented with erythromycin at 32°C was used to inoculate a fresh culture which was incubated at 37°C for five serial passages. The culture was diluted and plated on MRS containing erythromycin at 37°C to obtain single colonies that were replica streaked onto plates containing MRS with erythromycin and MRS with chloramphenicol to identify a chloramphenicol-sensitive, erythromycin-resistant clone.
Preparation of the bacterial strains and administration to mice.
The strains labeled with antibiotic resistance markers were grown overnight, harvested by centrifugation, washed with fresh media, and titrated to obtain a concentration of 109 CFU/ml. When these strains were inoculated into axenic mice, which did not receive antibiotics in their drinking water, no reversion (<1%) to antibiotic sensitivity was seen over a 14-day observation period. All experiments employing mice were performed using protocols approved by the Ethical Committee of the Canton de Vaud. Conventional female C3H/HeJ mice (Nestlé Research Centre, Lausanne, Switzerland) whose average age was 8 weeks were placed in Macrolon cages (five mice per cage) and housed in a room with a cycle consisting of 12 h of darkness and 12 h of light at 22°C. All mice had access to a standard diet (UAR 03-40; SAFE, Villemoisson, France) and water ad libitum.
For expression microarray analysis, the cages of the axenic mice were arranged in sterile isolators equipped with a sterile water supply. All mice were given access to irradiated standard chow diet (UAR 03-40; SAFE, Villemoisson, France) and autoclaved Vittel water. Then L. johnsonii NCC533 (Emr) was administered by intragastric gavage at a dose of 109 CFU to a group of five animals for three successive days.
Gut persistence experiments.
The colonization dynamics of strains was determined by culturing fecal pellets. L. lactis, E. coli, L. johnsonii, or 1:1 mixtures of mutant and parental L. johnsonii strains were administered by intragastric force feeding to groups of five animals for three successive days. Fecal samples were collected daily, weighed, and then diluted with 0.5 ml of Ringer's solution and mechanically homogenized. Dilutions were plated in triplicate on selective media and incubated at 37°C (or 30°C for L. lactis) for 2 days before enumeration. Dilutions were plated on selective media with chloramphenicol or erythromycin and incubated at 37°C for 2 days before enumeration. The plating was repeated twice for each fecal sample. Alternatively, the normal intestinal flora consisting mainly of Lactobacillus was monitored using MRS plates. No spontaneous Cmr or Emr Lactobacillus colonies were observed. The values described below are means and standard errors of the means for five biological replicates.
Microarray design.
DNA-based arrays were produced by Eurogentec S.A. (Liege, Belgium). The NCC533 microarrays covered 96% of the open reading frames (ORFs) of the complete genome. PCR amplicons that were 127 to 800 bp long corresponding to 1,756 ORFs identified from the L. johnsonii NCC533 genome sequence (3, 33) were spotted in duplicate onto slides (www.eurogentec.com). Amplicons from the luciferase gene (pSP-luc+) (www.promega.com) were also spotted as a spiking control. We used the same microarray platform for both genotyping and the expression array.
Comparative genomic hybridization.
Genomic DNA preparation and hybridization were performed as previously described (3), and the procedure included an additional purification step with Qiagen Genomic-tip 20/G (www.qiagen.com). Five nanograms of the pSP-luc+ plasmid was added to 1 μg of genomic DNA and labeled with FluoroLink Cy5- or Cy3-dUTP (www.amersham.com) using a DNA High Prime kit (www.roche-applied-science.com) according to the supplier's instructions. Hybridization was performed overnight at 42°C. Fluorescence scanning was performed with a ScanArray 4000 confocal laser scanner (www.perkinelmer.com). Signal intensities for each spot were determined using the Imagene software (www.biodiscovery.com). To preserve the signal intensity differences, we introduced into the labeling reaction mixtures a spiking control (pSP-luc+) and used its signal ratio to correct for differences in the labeling reaction efficiency. The signal ratio (ratio of Cy5-labeled unknown strain DNA to Cy3-labeled NCC533 DNA) of each spot was normalized using this spiking signal ratio as a reference.
Comparative genomic hybridization was performed in triplicate, with duplicate spots on each slide. The log2 signal ratio of each spot was then modified in order to shift the mean of the main peak of each hybridization reaction to this value. Finally, for each amplicon we obtained the mean of the ratios and the corresponding standard deviation. A gene corresponding to an amplicon was considered present in the unknown genome when the comparative genomic hybridization score of the mean of the ratios minus the standard deviation was greater than 200.
RNA isolation and amplification.
Axenic mice were maintained in sterile isolators. One week after monocolonization with a specified bacterial strain, groups of five mice were sacrificed using 3% isoflurane. The jejunum was dissected and washed with phosphate-buffered saline to remove the luminal contents. The mucosa-associated bacteria were recovered by scraping the mucosa and were flash-frozen in liquid nitrogen. The frozen tissue was crushed and suspended in equal volumes of Tris-EDTA buffer (pH 8) and phenol (pH 4.2). Total RNA was extracted by the Macaloid method as previously described (9). The cells were disrupted at maximum speed using a Mini-Bead Beater-8 (BioSpec Products, Bartlesville, OK) at 4°C; three 1-min cycles with a 1-min cooling period on ice between cycles were used. RNA was purified by phenol-chloroform extraction, followed by ethanol precipitation. Pellets were suspended in nuclease-free water, and 100 μg of total RNA was treated with 200 U of DNase I (Ambion, Huntingdon, United Kingdom) for 2 h at 37°C to eliminate contaminating DNA. An RNeasy mini kit (Qiagen, Basel, Switzerland) that was used for further purification included an additional on-column DNase digestion. RNA concentrations were determined spectrophotometrically. RNA of L. johnsonii obtained from tissue-scrapping samples was extracted from 100 μg of a mouse-bacterium RNA mixture using a MICROBEnrich kit (Ambion, Huntingdon, United Kingdom). For all samples the RNA integrity was tested using the Agilent RNA 6000 Nano assay (Agilent, Waldbronn, Germany).
cDNA synthesis, array hybridization, and analysis.
Total amplified RNA was hybridized to DNA arrays together with RNA extracted from mid-exponential-phase cells. For each hybridization, 2 μg of total RNA was labeled using a 3DNA Array 350RP Genisphere kit (Genisphere Inc., Hatfield, PA), as follows. Two micrograms of total RNA was preheated with random primers, kanamycin, and luciferase RNA. The reverse transcription reaction was performed with Superscript enzyme in its buffer supplemented with SUPERase-In for 2 h at 42°C. The reaction was stopped by addition of 0.5 M NaOH-50 mM EDTA and heating at 65°C for 15 min (protocol provided by the supplier). Spot signal intensities for each channel were corrected by subtracting the corresponding local background values. Spots displaying low intensities (i.e., intensities less than three times the standard deviation of the local background signal) were excluded. For all technical replicates, a control for fluorescence labeling by dye swapping was included. As the method is based on qualitative detection, negative and positive controls were used to confirm the absence of cross-hybridization and/or nonspecific hybridization. Many previously published microarray analyses expressed the in vitro transcription profile quantitatively by reference to mid-exponential broth growth. In the present study, we considered a gene expressed when its signal exceeded the background standard deviation by threefold. This procedure allowed comparison of the different expression patterns, but it yielded only qualitative data. To avoid a gene expressed by the autochthonous microflora, spots displaying a detectable signal with total RNA extracted from mice to which L. johnsonii NCC533 was not given orally were excluded from the in vivo analysis (less than 20 spots representing mainly ribosomal proteins were detected). The seemingly low in vivo expression level of isolate NCC533 reflects the strict selection criteria. A gene was considered “expressed” when it was detected in five or six experiments (three experiments with different mice, each performed in duplicate).
IgA protease assays.
IgA1-cleaving activity was detected by immunoelectrophoresis analysis of human and rat IgA, IgM, and IgG incubated overnight at 37°C with L. johnsonii wild-type strain NCC533, with a mutant with LJ1680 deleted, with Streptococcus mitis (positive control), or with sterile MRS in MEA medium (30). The samples were centrifuged, and each supernatant and pellet were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE). For detection of IgA1 protease activity after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, inhibition of sodium dodecyl sulfate was overcome and proteins were renatured by incubation for 2 h at room temperature in 50 mM Tris-HCl (pH 7.4)-0.154 M NaCl containing 1% Triton X-100 and 0.5% bovine serum albumin. The membrane was rinsed in alcohol and then in water and incubated for 1 h with rabbit antibody directed against human and rat Igs. The proteins in the gel were allowed to react with IgA1 on the membrane for 20 to 60 min at 37°C in a humid chamber. After this the membrane was washed, and cleavage of IgA1 was visualized by incubation with peroxidase-conjugated rabbit antibody. In this assay IgA1 protease activity in the gel was reflected by a loss of light chains (as part of Fab fragments) and was detected on the membrane as a lack of staining on a red background.
Sugar fermentation.
API 50 CH (bioMérieux) carbohydrate fermentation strips and API 50 CHL medium were used according to the instructions of the supplier.
RESULTS
In vitro growth and gut persistence.
The intestinal L. johnsonii isolate NCC533 and the L. johnsonii type strain, the blood isolate ATCC 33200T, showed unrelated restriction patterns (50). Their growth rates (doubling times, 41.3 ± 0.5 and 39.8 ± 0.9 min, respectively) and the final pH values (pH 3.68 ± 0.1 and 3.81 ± 0.03, respectively) in MRS broth cultures were indistinguishable (means ± standard errors of the means of three experiments).
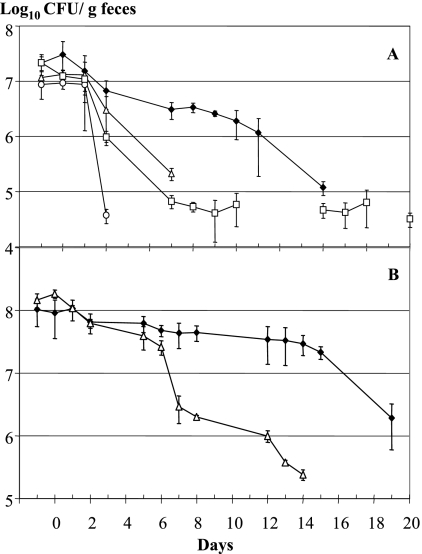
Over 3 days groups of five conventional mice received a single daily dose of 108 CFU of isolate NCC533 or strain ATCC 33200T by intragastric force feeding. In mice fed isolate NCC533, fecal titers greater than 106 CFU per g of feces were maintained over 8 days, while mice given strain ATCC 33200T maintained the inoculated bacterium at this titer for only 1 day (Fig. 1A). Fecal isolate NCC533 and strain ATCC 33200T remained detectable for 12 and 5 days, respectively. L. lactis, a control for passive gut transit by a dairy lactic acid bacterium (LAB) (10, 16), was quickly eliminated from the gut (Fig. 1A). E. coli strain K-12, isolated from a human stool (41), was detected, although the fecal titers were low during the experiment (Fig. 1A).
FIG. 1.
L. johnsonii strains with different isolation histories have distinct gut residence times. The stool-derived L. johnsonii strain NCC533 (Emr) (⧫) and the blood-derived L. johnsonii type strain ATCC 33200T (Cmr) (▵) differ significantly in gut persistence in untreated (A) or antibiotic-treated (B) conventional mice. The fecal colony counts of the bacteria are plotted as log10 titers against the time (in days) after 3 days of force feeding with the corresponding bacteria. For comparison, another gut commensal, E. coli strain K-12 (Ampr) (□), and the dairy bacterium L. lactis strain MG1363 (Cmr) (○) were given to untreated conventional mice (A). The symbols indicate the arithmetic means, and the error bars indicate the standard errors of the means for groups of five mice.
The feeding experiment was repeated with mice that were fed antibiotics. This treatment reduced the endogenous gut Lactobacillus count from 109 to 104 CFU/g feces. The fecal titers were higher for isolate NCC533 and strain ATCC 33200T, and the gut persistence time was prolonged slightly for both test strains, but the gut persistence times of the two strains again were clearly different (Fig. 1B).
Genotyping microarray analysis revealed isolate NCC533-specific genes.
To identify genes associated with the long-gut-persistence phenotype, we first searched for genes in isolate NCC533 which lacked a counterpart in strain ATCC 33200T. Strain ATCC 33200T DNA did not hybridize with 233 of the 1,756 isolate NCC533 ORFs spotted on the microarray. Most isolate NCC533-specific genes represented clusters of adjacent genes (Fig. 2; see Table S3 in the supplemental material), and 30% were prophage related. Other isolate NCC533-specific gene clusters included a chemical resistance cassette and regions encoding surface proteins and translocases, PTS transporters, bacteriocin, and proteins involved in exopolysaccharide (EPS) synthesis. Some genetic differences between isolate NCC533 and strain ATCC 33200T were also represented by isolated NCC533-specific genes (see Table S3 in the supplemental material). We do not want to imply that the differences between the two investigated isolates represent functions relevant for niche-specific fitness (i.e., gut adaptation versus blood adaptation).
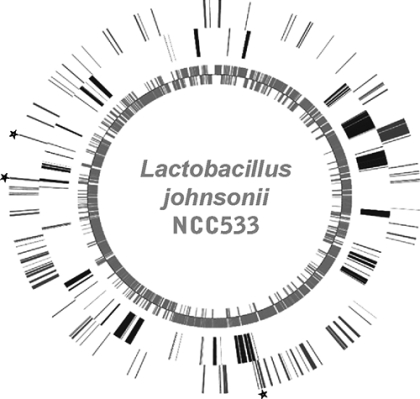
FIG. 2.
Results of dual microarrays projected on the genome map of the gut-adapted L. johnsonii strain NCC533. The genome organization is indicated, with counterclockwise- and clockwise-oriented ORFs shown in the innermost circle and the next circle, respectively. The next circle indicates the 233 ORFs in the reference strain that did not hybridize with the genomic DNA from the blood-derived reference strain. The outermost circle indicates the 174 genes from the gut-adapted strain that were expressed in bacteria associated with jejunal mucosa from monocolonized mice. Asterisks indicate ORFs that were NCC533 specific and in vivo expressed.
In vivo expression microarray analysis.
Gene content analysis of the two strains yielded too many strain-specific genes, and the data therefore had to be combined with other data to associate the observed phenotype with a genotype. To achieve this goal, we isolated mRNA from isolate NCC533 associated with the jejunal mucosa in monocolonized mice. A total of 174 isolate NCC533 ORFs were expressed in this intestinal compartment. The list of the jejunum-expressed isolate NCC533 genes revealed genes involved in basic cellular anabolic activities like protein, DNA, RNA, cell wall, and EPS synthesis (see Table S4 in the supplemental material). On the catabolic side, genes encoding the import and degradation of sugars via the glycolytic pathway were prominent.
Fusion of the two microarray data sets.
When the results of strain ATCC 33200T-isolate NCC533 comparative genomic hybridization and the isolate NCC533 gene expression microarray analysis for monocolonized mice were combined, we observed that only six isolate NCC533-specific genes, corresponding to three genetic loci, were transcribed in bacteria from the jejunal mucosa of monocolonized mice (Fig. 2). The first locus was LJ1027-LJ1028, encoding two glycosyltransferase genes in the EPS synthesis cluster (see Table S5 in the supplemental material). The second locus was LJ1654 to LJ1656. LJ1654 encodes membrane-bound transporter protein subunit C for a phosphoenolpyruvate-dependent sugar PTS (annotated as the mannose PTS). LJ1655 and LJ1656 encode the dissociable cytoplasmic AB protein complex of this sugar import complex. The third gut-expressed and isolate NCC533-specific locus was LJ1680, which shared significant sequence identity (30% amino acid identity; E value, 10−80) with IgA proteases from several pathogenic bacteria that colonize human body surfaces (including Streptococcus pyogenes and Staphylococcus aureus).
In vitro phenotype of the KO mutants.
We constructed KO mutants of isolate NCC533 by allelic replacement in the three loci using an erythromycin resistance cassette. Antibiotic-resistant isogenic mutants were obtained, verified by PCR, and investigated to determine whether they had a phenotype that could be observed in vitro. Notably, the in vitro growth kinetics of the mutant and parental NCC533 strains in MRS broth lacking antibiotics did not differ. Furthermore, the cell counts for the mutant and the parental NCC533 strain were identical when early-stationary-phase growth in MRS broth was assessed.
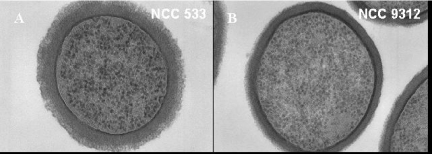
For the glycosyltransferase genes, we constructed a KO mutant that removed the entire EPS gene cluster, namely, LJ1021 to LJ1035 (3). Compared to the parental strain (Fig. 3A), the isogenic KO mutant lacked the fuzzy layer decorating the outer rim of the cell wall as determined by electron microscopy observation (Fig. 3B), as described previously for nonencapsulated pathogenic streptococci (2). With the putative mannose PTS mutant, we observed a distinct sugar growth pattern when a commercial API 50 gallery was used (data not shown). However, both the parental and mutant strains could grow on mannose. Notably, isolate NCC533 contains two additional mannose PTS clusters, and their expression was detected in vitro but not in vivo. For the putative IgA protease mutant, we observed that both the mutant and the wild-type strain showed marked proteolytic activity with Igs added to conditioned growth medium. The activity was not specific for the isotype (IgA or IgG) or species (mouse or human) of the antibodies. The proteolytic activity of L. johnsonii was much stronger than that of an S. pyogenes strain with a known IgA protease activity (data not shown).
FIG. 3.
Ultrathin section of the parental L. johnsonii strain NCC533 (A) and the mutant NCC533 ΔLJ_1021-1035::Emr (B) with the entire eps gene cluster deleted. Note the fuzzy layer on the outside of the cell wall of the wild-type strain, which is not present in the KO mutant.
Test KO mutants.
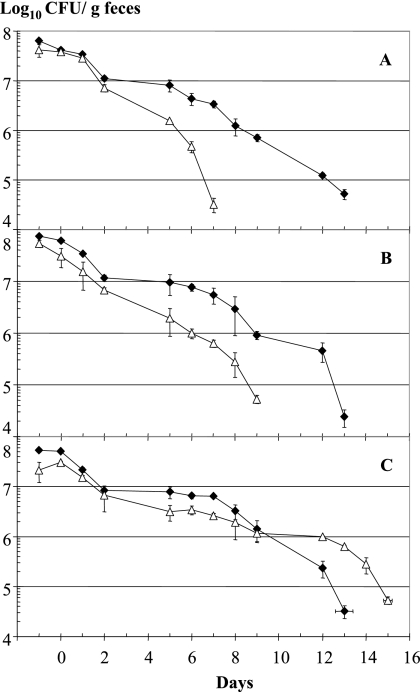
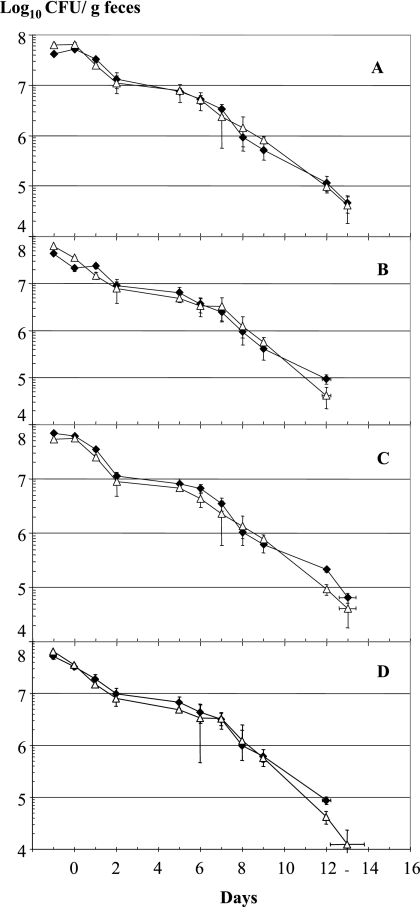
Deletion of LJ1654 to LJ1656 (PTS transporter KO) and LJ1680 (IgA protease KO) resulted in a significantly reduced persistence time for the mutant strain in the mouse intestine (Fig. 4A and B) (groups of five animals). In contrast, deletion of the entire EPS cluster did not decrease but slightly increased the gut persistence time of the KO mutant compared to the parental strain (Fig. 4C).
FIG. 4.
Gut persistence times for three mutants carrying deletions in the three genetic loci associated with the phenotype determined by dual microarray analysis. Competition experiments were performed with the wild-type (Cmr) strain (⧫) and array-predicted mutant strains (▵). The mutant strains were (A) NCC533 ΔLJ_1654-1056::Emr (PTS sugar transporter), (B) NCC533 ΔLJ_1680::Emr (IgA protease), and (C) NCC533 ΔLJ_1021-1035::Emr (eps gene cluster). For further details see the legend to Fig. 1.
Control KO mutants.
As a negative control, we first verified that the replacement technology had no negative effect on the gut persistence time of the mutant strain. To do this, we inactivated LJ0667, which encodes a large membrane-inserted protein having 10 transmembrane domains and 14 leucine zippers. No effect on gut persistence was observed with this mutant (Fig. 5C). In a second control experiment, we inactivated LJ1840, which encodes the protease PrtP. This gene is one of the isolate NCC533-specific ORFs that were not transcribed in vivo. Dairy LAB depend on similar cell wall-bound proteinases for the degradation of and growth on milk proteins (27). Notably, the LJ1840 KO mutant behaved like the wild-type strain in the gut persistence assay (Fig. 5A).
FIG. 5.
Gut persistence times for control KO mutants. Competition experiments were performed with the parental wild-type strain (Cmr) (⧫) and four KO deletion mutants (▵). The mutant strains were (A) NCC533 ΔLJ_1840::Emr (cell wall protease PrtP), (B) NCC533 ΔLJ_1476::Emr (sortase), (C) NCC533 ΔLJ_0667::Emr (anonymous membrane protein), and (D) NCC533 ΔLJ_1412/ΔLJ_1147/ΔLJ_0056::Emr (three bile salt hydrolases). For further details see the legend to Fig. 1.
A KO of a large surface protein affected the gut performance of L. reuteri in mice (51). Isolate NCC533 contains a homologue of this protein. To target cell wall-anchored surface protein genes collectively, we knocked out the transpeptidase sortase gene LJ1476, which encodes an enzyme required for the anchoring of LPXTG-containing proteins to the cell wall (24, 43). Notably, the LJ1476 KO mutant showed a gut persistence pattern identical to that of the wild-type strain (Fig. 5B).
A number of in vitro experiments have indicated that bile resistance is a crucial factor for gut persistence of commensals and that several intestinal bifidobacteria and lactobacilli encode bile salt hydrolases (6, 33, 39). We created a triple KO mutant for the bile salt hydrolase proteins encoded by the LJ0056, LJ1147, and LJ1412 genes of isolate NCC533. Again, the triple KO mutant showed a gut persistence pattern indistinguishable from that of the parental strain (Fig. 5D). These negative results underline the value of the positive results obtained with two (PTS transporter and IgA protease) of the three gene clusters identified in our screening analysis.
DISCUSSION
The rapid gut transit of an L. lactis strain demonstrates that a dairy strain is quickly expelled from the mouse intestine by peristalsis. L. lactis probably travels with the food bolus. This is not the case for the investigated Lactobacillus strains.
In the present study we used a combination of genotyping and expression microarrays to investigate the genetic basis for a phenotypic difference between two L. johnsonii strains with different gut persistence times. The two strains contained different antibiotic resistance markers to facilitate enumeration of the bacteria. As we lacked sequence information for strain ATCC 33200T to perform a strain ATCC 33200T-specific PCR test, we could not differentiate the two test strains from each other and from the endogenous murine lactobacilli using quantitative PCR tests. Isolate NCC533 contained a chromosomally integrated erythromycin resistance gene, while strain ATCC 33200T contained a plasmid-encoded Cmr gene integrated into the chromosome via the integrase of prophage Lj771 (E. Denou, R. D. Pridmore, M. Ventura, A. C. Pittet, M. C. Zwahlen, B. Berger, C. Barretto, J. M. Panoff, and H. Brüssow, submitted for publication). Theoretically, the distinct integration system could lead to a decrease in the growth rate of strain ATCC 33200T or decreased stability of the antibiotic resistance marker in strain ATCC 33200T compared to isolate NCC533. Both possibilities could undermine the hypothesis that strain ATCC 33200T lacks important colonization determinants which isolate NCC533 possesses. A number of experimental observations argue against this concern. First, during growth in broth strain ATCC 33200T-Cmr and isolate NCC533-Emr showed identical in vitro growth properties when they were maintained in the absence of antibiotic selection, indicating that the Cmr construction does not have a growth-retarding effect. Second, with the same cellular background isolates NCC533-Emr and NCC533-Cmr showed identical in vivo persistence times in mice that were or were not treated with antibiotics. Third, when both antibiotic-resistant strains were cocultivated starting with equal cell numbers, identical ratios of the strains were observed after a 24-h growth period. This technique is quite sensitive for revealing even subtle differences in the growth rate. Fourth, we explored the stability of the erythromycin resistance gene. After 20 serial passages in the absence of antibiotics, all 200 colonies tested still contained the marker gene when they were tested on culture plates lacking antibiotics and on culture plates containing antibiotics. Likewise, when given to mice in vivo all erythromycin-resistant colonies were identified by PCR as colonies of isolate NCC533 strains. Notably, no transfer of the resistance gene to endogenous lactobacilli was observed. Finally, the chromosomal integration via the Lj771 prophage integrase is stable in L. johnsonii (Denou et al., submitted).
Taken together, these observations indicate that the resistance marker does not have an influence on the persistence time, and we concluded that the colonization determinants of isolate NCC533 and strain ATCC 33200T genuinely differ. We then hypothesized that genes responsible for the “long-gut-persistence” phenotype must be specific to the strain having the phenotype and have to be expressed in the intestine. When we fused the two microarray data sets, only six genes at three loci fulfilled these criteria for strain NCC533. We did not expect a large number of candidate genes since expression profiling reveals mainly housekeeping activities that are widely shared by different L. johnsonii strains (3, 9), while the strain-specific DNA represents much of the selfish DNA that for the most part is transcriptionally silent (9; Denou et al., submitted). However, we stress that our approach probably underestimates the number of genes affecting the long-gut persistence phenotype. Using expression data from other gut segments (e.g., stomach), other gut locations (luminal bacteria versus mucosa-associated bacteria), or different animals (axenic mice versus conventional mice) (9) might lead to the identification of further genes that affect the gut persistence phenotype. It is likely that the gut persistence phenotype is affected by many genes. Despite this caveat, we confirmed with KO mutants that two of the three genetic loci identified in the present study affected the gut residence time. The combination of genotyping and expression microarray techniques is thus a promising approach for defining the genetic basis of complex in vivo bacterial phenotypes.
What did we learn from the bioinformatic annotation and the phenotype of the mutants about the physiological basis of gut persistence? One locus identified in our screen is annotated as a sugar import system, namely, a PTS transporter. This annotation was confirmed by the distinct sugar utilization pattern of the KO mutant compared to the parental strain. These observations indicate that sugar import is a crucial and specific factor affecting the gut persistence of isolate NCC533. This is a plausible conclusion, which concurs with the IVET data for L. plantarum identifying PTS genes as prominent in vivo-expressed genes and the data for KO mutants (7). Genome analysis of dominant gut bacteria like Bacteroides and Bifidobacterium has underlined the importance of sugar catabolism for gut commensalism (55). The LJ1654 to LJ1656 genes are annotated as genes encoding a mannose-specific PTS. However, the mannose utilization of the parental and mutant strains did not differ (utilization was positive in both cases). In fact, one might question whether a mannose fermentation defect could have been observed since the isolate NCC533 genome contains three other loci annotated as genes encoding mannose-specific PTS transporters (LJ0503 to LJ0505, LJ0627 to LJ0629, and LJ0739 to LJ0742). The LJ0503 to LJ0505 genes are in addition expressed in the small intestine, but since these genes are shared by isolate NCC533 and strain ATCC 33200T, they were not identified in our screen.
The second gene that decreased the gut persistence time in L. johnsonii is annotated as an IgA protease gene. Numerous reports have suggested that the immune system controls the composition of the gut microbiota (12, 20, 21, 40). Indeed, the variable surface makeup of intestinal Bacteroides species suggests independently that even major gut commensals are under immune selection (8, 17). There are data that specifically link the presence of IgA protease with adsorption of pathogenic LAB on mucosal sites. Expression of an IgA1 protease by Streptococcus pneumoniae cleaves the antibody in the hinge region such that only the Fab fragment binds the bacterial cell, which increases the binding of the bacteria to the respiratory mucosa (53). The bound Fab fragment is hydrophobic and allows better binding of the streptococcal cell. However, one should not prematurely embrace these appealing parallels. No IgA-specific proteolytic activity was observed in our test strain, and its generalized proteolytic activity (L. johnsonii depends on amino acid import for growth [46]) was not affected by the KO of LJ1680. Thus, the role of this gene remains to be defined; apparently the 30% amino acid sequence identity with proven IgA proteases is too low to predict a function.
Finally, the identification of the eps genes in our dual microarray screen is problematic. The dilemma is not the observation that the eps KO mutant had a slightly increased gut persistence time instead of a decreased persistence time. Actually, increased persistence of the eps KO mutant would even be expected. Since the bacterial capsular polysaccharides are negatively charged, they electrostatically interfere with the binding to the receptor on the mucosal membrane. The loss of the capsular polysaccharide layer should thus lead to increased gut persistence of the mutant NCC533 strain. In fact, S. pneumoniae strains lacking the capsular polysaccharide layer also showed increased adherence to target cells (14, 35, 53) compared to isogenic but capsulated strains. Furthermore, in some bacteria the capsule can shield the function of short bacterial adhesins (37). Our concern with the identification of the eps genes in our screen is technical. We know from comparative sequencing that the eps cluster is a region with high genetic diversity in L. johnsonii (3) and that it is strongly expressed in a variety of growth conditions (9). Genes with such properties are automatically selected in the proposed genotyping-expression microarray test without being causally linked to the investigated phenotype, leading to false-positive associations.
Finally we stress that gut persistence is necessary but not sufficient for an organism to have probiotic effects. It will therefore be important to apply the described combination of genotyping and expression microarray analyses to a medically relevant phenotype of probiotic bacteria. We are currently testing this approach with the antidiarrheal disease phenotype displayed by some probiotic bacteria in a mouse rotavirus infection model (26).
Supplementary Material
Acknowledgments
We thank Marie-Camille Zwahlen and Caroline Barretto for help with the bioinformatic analyses, Marie-Lise Dillmann for electron microscopy, Catherine Schwartz and Massimo Marchesini for assistance with the animal experiments, and Anne-Cécile Pittet for technical assistance. We also thank Annick Mercenier for comments and fruitful discussions.
Footnotes
Published ahead of print on 25 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, S. J., B. Okoko, E. Martinez, G. Gregorio, and L. F. Dans. 2004. Probiotics for treatment of infectious diarrhoea. Cochrane Database Syst. Rev. 2CD003048. [DOI] [PubMed] [Google Scholar]
- 2.Benga, L., R. Goethe, M. Rohde, and P. Valentin-Weigand. 2004. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell. Microbiol. 6867-881. [DOI] [PubMed] [Google Scholar]
- 3.Berger, B., R. D. Pridmore, C. Barretto, F. Delmas-Julien, K. Schreiber, F. Arigoni, and H. Brüssow. 2007. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 1891311-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergonzelli, G. E., S. Blum, H. Brüssow, and I. Corthesy-Theulaz. 2005. Probiotics as a treatment strategy for gastrointestinal diseases? Digestion 7257-68. [DOI] [PubMed] [Google Scholar]
- 5.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 1865721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 1867829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bron, P. A., M. Meijer, R. S. Bongers, W. M. de Vos, and M. Kleerebezem. 2007. Dynamics of competitive population abundance of Lactobacillus plantarum ivi gene mutants in faecal samples after passage through the gastrointestinal tract of mice. J. Appl. Microbiol. 1031424-1434. [DOI] [PubMed] [Google Scholar]
- 8.Cerdeno-Tarraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 3071463-1465. [DOI] [PubMed] [Google Scholar]
- 9.Denou, E., B. Berger, C. Barretto, J. M. Panoff, F. Arigoni, and H. Brüssow. 2007. Gene expression of commensal Lactobacillus johnsonii strain NCC533 during in vitro growth and in the murine gut. J. Bacteriol. 1898109-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drouault, S., G. Corthier, S. D. Ehrlich, and P. Renault. 1999. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 654881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne, C., L. O'Mahony, L. Murphy, G. Thornton, D. Morrissey, S. O'Halloran, M. Feeney, S. Flynn, G. Fitzgerald, C. Daly, B. Kiely, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 2001. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73386S-392S. [DOI] [PubMed] [Google Scholar]
- 12.Fagarasan, S., M. Muramatsu, K. Suzuki, H. Nagaoka, H. Hiai, and T. Honjo. 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 2981424-1427. [DOI] [PubMed] [Google Scholar]
- 13.Fukushima, Y., S. Miyaguchi, T. Yamano, T. Kaburagi, H. Iino, K. Ushida, and K. Sato. 2007. Improvement of nutritional status and incidence of infection in hospitalised, enterally fed elderly by feeding of fermented milk containing probiotic Lactobacillus johnsonii La-1 (NCC533). Br. J. Nutr. 98969-977. [DOI] [PubMed] [Google Scholar]
- 14.Hulse, M. L., S. Smith, E. Y. Chi, A. Pham, and C. E. Rubens. 1993. Effect of type III group B streptococcal capsular polysaccharide on invasion of respiratory epithelial cells. Infect. Immun. 614835-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humen, M. A., G. L. de Antoni, J. Benyacoub, M. E. Costas, M. I. Cardozo, L. Kozubsky, K. Y. Saudan, A. Boenzli-Bruand, S. Blum, E. J. Schiffrin, and P. F. Perez. 2005. Lactobacillus johnsonii La-1 antagonizes Giardia intestinalis in vivo. Infect. Immun. 731265-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimoto, H., M. Nomura, M. Kobayashi, K. Mizumachi, and T. Okamoto. 2003. Survival of lactococci during passage through mouse digestive tract. Can. J. Microbiol. 49707-711. [DOI] [PubMed] [Google Scholar]
- 17.Krinos, C. M., M. J. Coyne, K. G. Weinacht, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414555-558. [DOI] [PubMed] [Google Scholar]
- 18.Leong-Morgenthaler, P., M. C. Zwahlen, and H. Hottinger. 1991. Lactose metabolism in Lactobacillus bulgaricus: analysis of the primary structure and expression of the genes involved. J. Bacteriol. 1731951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lievin-Le Moal, V., and A. L. Servin. 2006. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 19315-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macpherson, A. J., D. Gatto, E. Sainsbury, G. R. Harriman, H. Hengartner, and R. M. Zinkernagel. 2000. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2882222-2226. [DOI] [PubMed] [Google Scholar]
- 21.Macpherson, A. J., and T. Uhr. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 3031662-1665. [DOI] [PubMed] [Google Scholar]
- 22.Macrina, F. L., J. A. Tobian, K. R. Jones, R. P. Evans, and D. B. Clewell. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19345-353. [DOI] [PubMed] [Google Scholar]
- 23.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285760-763. [DOI] [PubMed] [Google Scholar]
- 25.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 1876119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pant, N., H. Marcotte, H. Brüssow, L. Svensson, and L. Hammarström. 2007. Effective prophylaxis against rotavirus diarrhea using a combination of Lactobacillus rhamnosus GG and antibodies. BMC Microbiol. 7e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastar, I., I. Tonic, N. Golic, M. Kojic, R. van Kranenburg, M. Kleerebezem, L. Topisirovic, and G. Jovanovic. 2003. Identification and genetic characterization of a novel proteinase, PrtR, from the human isolate Lactobacillus rhamnosus BGT10. Appl. Environ. Microbiol. 695802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavan, S., P. Desreumaux, and A. Mercenier. 2003. Use of mouse models to evaluate the persistence, safety, and immune modulation capacities of lactic acid bacteria. Clin. Diagn. Lab. Immunol. 10696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulsen, K., J. Reinholdt, and M. Kilian. 1996. Characterization of the Streptococcus pneumoniae immunoglobulin A1 protease gene (iga) and its translation product. Infect. Immun. 643957-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pretzer, G., J. Snel, D. Molenaar, A. Wiersma, P. A. Bron, J. Lambert, W. M. de Vos, R. van der Meer, M. A. Smits, and M. Kleerebezem. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 1876128-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56309-312. [DOI] [PubMed] [Google Scholar]
- 33.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 1012512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rembacken, B. J., A. M. Snelling, P. M. Hawkey, D. M. Chalmers, and A. T. Axon. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354635-639. [DOI] [PubMed] [Google Scholar]
- 35.Ring, A., J. N. Weiser, and E. I. Tuomanen. 1998. Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Investig. 102347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saavedra, J. M., N. A. Bauman, I. Oung, J. A. Perman, and R. H. Yolken. 1994. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet 3441046-1049. [DOI] [PubMed] [Google Scholar]
- 37.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Servin, A. L. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28405-440. [DOI] [PubMed] [Google Scholar]
- 39.Sheehan, V. M., R. D. Sleator, C. Hill, and G. F. Fitzgerald. 2007. Improving gastric transit, gastrointestinal persistence and therapeutic efficacy of the probiotic strain Bifidobacterium breve UCC2003. Microbiology 1533563-3571. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki, K., B. Meek, Y. Doi, M. Muramatsu, T. Chiba, T. Honjo, and S. Fagarasan. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 1011981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweeney, N. J., D. C. Laux, and P. S. Cohen. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 643504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tannock, G. W. 2004. A special fondness for lactobacilli. Appl. Environ. Microbiol. 703189-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 9612424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuomola, E., R. Crittenden, M. Playne, E. Isolauri, and S. Salminen. 2001. Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr. 73393S-398S. [DOI] [PubMed] [Google Scholar]
- 45.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 4441027-1031. [DOI] [PubMed] [Google Scholar]
- 46.van der Kaaij, H., F. Desiere, B. Mollet, and J. E. Germond. 2004. l-Alanine auxotrophy of Lactobacillus johnsonii as demonstrated by physiological, genomic, and gene complementation approaches. Appl. Environ. Microbiol. 701869-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Niel, C. W., C. Feudtner, M. M. Garrison, and D. A. Christakis. 2002. Lactobacillus therapy for acute infectious diarrhea in children: a meta-analysis. Pediatrics 109678-684. [DOI] [PubMed] [Google Scholar]
- 48.Vaughan, E. E., R. D. Pridmore, and B. Mollet. 1998. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 1804893-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura, M., C. Canchaya, D. Pridmore, B. Berger, and H. Brüssow. 2003. Integration and distribution of Lactobacillus johnsonii prophages. J. Bacteriol. 1854603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura, M., and R. Zink. 2002. Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 217141-154. [DOI] [PubMed] [Google Scholar]
- 51.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter, J., N. C. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 692044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiser, J. N., D. Bae, C. Fasching, R. W. Scamurra, A. J. Ratner, and E. N. Janoff. 2003. Antibody-enhanced pneumococcal adherence requires IgA1 protease. Proc. Natl. Acad. Sci. USA 1004215-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, M. 2005. Microbial inhabitants of humans. Their ecology and role in health and disease. Cambridge University Press, Cambridge, United Kingdom.
- 55.Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 10010452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.