Abstract
We report the discovery and molecular characterization of a small and very acidic nucleolar protein of an SDS/PAGE mobility corresponding to Mr 29,000 (NO29). The cDNA-deduced sequence of the Xenopus laevis protein defines a polypeptide of a calculated molecular mass of 20,121 and a pI of 3.75, with an extended acidic region near its C terminus, and is related to the major nucleolar protein, NO38, and the histone-binding protein, nucleoplasmin. This member of the nucleoplasmin family of proteins was immunolocalized to nucleoli in Xenopus oocytes and diverse somatic cells. Protein NO29 is associated with nuclear particles from Xenopus oocytes, partly complexed with protein NO38, and occurs in preribosomes but not in mature ribosomes. The location and the enormously high content of negatively charged amino acids lead to the hypothesis that NO29 might be involved in the nuclear and nucleolar accumulation of ribosomal proteins and the coordinated assembly of pre-ribosomal particles.
The packaging of DNA and histones into nucleosomes as a first step in chromatin formation and the packaging of rRNA and ribosomal proteins (r-proteins) during ribosome formation are, at first glance, two basically different and for the most part spatially separated processes. The former occurs at chromatin borders in the nucleoplasm and the latter is confined to the nucleolus. However, in both processes, the eukaryotic cell has to overcome a common chemical problem—i.e., the prevention of nonspecific associations and aggregations of these basic proteins with the numerous negatively charged nucleic acids and proteins abundant in the cytoplasm and in the nucleus. Biochemical studies have identified several proteins containing acidic regions that bind histones and can transfer them onto DNA to form nucleosome cores. Examples include major proteins of the amphibian oocyte such as nucleoplasmin and protein N1/N2 (1–5) as well as somatic cell proteins such as “nucleoplasmin S” (6), CAF-1 (7), and other related proteins (8–10).
During ribosome assembly, the concerted interaction of r-proteins and rRNA is believed to involve certain nonribosomal nucleolar proteins (reviewed in 11–13). One of the best characterized among them is protein NO38, a major constituent of the granular component of nucleoli of Xenopus laevis (14), a homolog of mammalian protein B23 (15–19). Like nucleoplasmin (20), protein NO38/B23 tends to form stable oligomers (14, 21) and can shuttle between the nucleus and the cytoplasm (22). Because of its sequence homology to nucleoplasmin and its reported ability to bind to nucleic acids (23–25), protein NO38/B23 has been proposed to promote ribosome assembly and transport across the nuclear envelope (12). A direct association of protein NO38/B23 and r-proteins, however, has not yet been demonstrated. In addition, protein NO38/B23 has been reported to possess ribonuclease activity (26) and to associate with transcription factor YY1 (27), protein Rev of HIV-1 (28–30), and nucleolar protein p120 (31), suggesting that this protein has multiple functions.
Although sequence elements required for the specific nucleolar accumulation of protein NO38/B23 have been described (32, 33), its direct and constitutive binding partners within the nucleolus remain to be identified. In immunoprecipitation experiments and affinity chromatography using cellular and nuclear extracts of X. laevis cultured cells and antibodies to protein NO38 (14), we have recently isolated and cDNA-cloned a novel nucleolar, very acidic protein with an SDS/PAGE mobility corresponding to Mr 29,000 (NO29), which reveals a striking amino acid homology to both nucleoplasmin and protein NO38.
MATERIALS AND METHODS
Biological Material.
Clawed toads (X. laevis) were purchased from the South African Snake Farm (Krysna, Republic of South Africa). Culture conditions for X. laevis kidney epithelial cells (XLKE, line A6) and human hepatocellular carcinoma cells of line PLC have been described (34).
Isolation and Fractionation of Xenopus Oocyte Nuclei, Egg Extracts, and Ribosomal Subunits.
Nuclei of mature (stages IV–VI) X. laevis oocytes were isolated and fractionated (35) into low speed pellet (LSP), high speed pellet (HSP), and high speed supernatant (HSS). LSP fractions were cleared from yolk proteins by Freon extraction (36). Preparations of total egg extracts (37), ribosomes, and ribosomal subunits have been described (38).
Immunoprecipitations from Cellular and Nuclear Extracts of A6 Cells.
For l-[35S]methionine labeling, A6 cells were grown in methionine-reduced minimal essential medium, with 10% fetal calf serum and l-[35S]methionine, for 16 h before harvest. Cellular extracts were prepared as described (33). Nuclei from somatic cells were prepared according to Miake-Lye and Kirschner (39), resuspended in 5:1 buffer (10 mM Tris⋅HCl, pH 7.4/83 mM KCl/l7 mM NaCl/2 mM MgCl2), homogenized by sonication (Branson sonifier B-12; 5 × 20 sec, 75 W), and fractionated as described for oocyte nuclei. The resulting LSP was extracted in PBS, supplemented with 250 mM NaCl, for 20 min on ice. In vivo-labeled cellular or nuclear extracts from A6 cells were used for immunoprecipitations with mAb No-185 or No-63 (33). For preparative immunochromatography of proteins from A6 cell extracts, mAb No-185 was covalently coupled to agarose beads using the Immunopure Ag/Ab Immobilization Kit #3 (Bender und Hobein, Bruchsal, Germany).
Antibodies and Antibody Purification.
The Xenopus-specific protein NO38 mAbs, No-185 and No-63, and the myc mAb 9E10 have been described (14, 33). Antibodies specific for the newly identified protein NO29 were obtained by immunization of guinea pigs with two synthesized peptides (40), EVTVPLANLK and SGFISSTAAQGPPSPAIE, coupled to keyhole limpet hemocyanin. While the first peptide sequence had originally been identified by direct amino acid sequencing of a polypeptide spot obtained after immunochromatography of cellular extracts with mAb No-185 and two-dimensional (2D) gel electrophoresis of the eluted proteins, the second one was deduced from the NO29 cDNA. All results shown have been obtained with antibodies affinity-purified on iodoacetyl-immobilized peptides (40).
Gel Electrophoresis and Immunoblotting.
SDS/PAGE and 2D-PAGE of proteins, using either isoelectric focusing (IEF) or nonequilibrium pH gradient electrophoresis (NEPHGE) in the first dimension separation were as described (14, 34, 35, 37). Immunoblotting was performed on Immobilon polyvinylidene difluoride (PVDF) membranes (Millipore) as described (33), using protein NO29 antibodies, diluted 1:500 in Tris-buffered saline, and peroxidase-coupled secondary antibodies (Dianova, Hamburg, Germany) detected with the ECL system (Amersham).
Isolation of cDNA Clones.
Total DNA from a λ Unizap cDNA library from X. laevis kidney (Stratagene) was used for PCR with the library-specific reverse primer as sense primer and a degenerated antisense primer deduced from the amino acid sequence DEHNVVEVTAPN. An amplified 249-nt cDNA fragment was used as “random primed”, 32P-labeled probe for screening the same cDNA library. One of 16 isolated positive clones, termed pBT-NO29, contained a full mRNA length cDNA and was used for further analyses.
RNA Isolation and Northern Blot Hybridization.
Total RNA from Xenopus ovaries or A6 cells was prepared as described (41, 42), and poly(A)+ RNA with the mRNA Purification Kit (Pharmacia). Northern blot analysis was performed as described (34) using a 32P-labeled antisense cRNA probe corresponding to nt 1–249 of clone pBT-NO29. Hybridization and washing were at 65°C.
In Vitro Synthesis of cDNA-Cloned Proteins.
Clone pBT-NO29 was transcribed and translated in vitro using a coupled reticulocyte (TnT) system (Promega). Immunoprecipitations were performed (33) using protein NO29 antibodies coupled to protein A-Sepharose (Pharmacia). Translation products and immunoprecipitates were analyzed by SDS/PAGE and autoradiography.
Transfection Experiments.
For transient expression in human PLC cells, protein NO29 cDNA was subcloned into the BT-myc vector (43), and the resulting construct further subcloned into the eukaryotic expression vector pRcCMV (ITC Biotechnology, Heidelberg). Transfections were carried out as described (33).
Immunofluorescence Microscopy.
Fixation and immunocytochemistry of transiently transfected PLC cells was as described (33). Cultured A6 cells were fixed with 2% formaldehyde for 20 min at room temperature (RT), followed by incubation in PBS containing 50 mM NH4Cl (3 min at RT), and finally in PBS containing 0.5% Triton X-100 (5 min at RT). After two washes in PBS (each at 5 min at RT), specimens were incubated with antibodies that had been diluted 1:50 in PBS for 20 min at RT.
RESULTS
Coimmunoprecipitations from Cellular and Nuclear Extracts Using mAbs to Protein NO38.
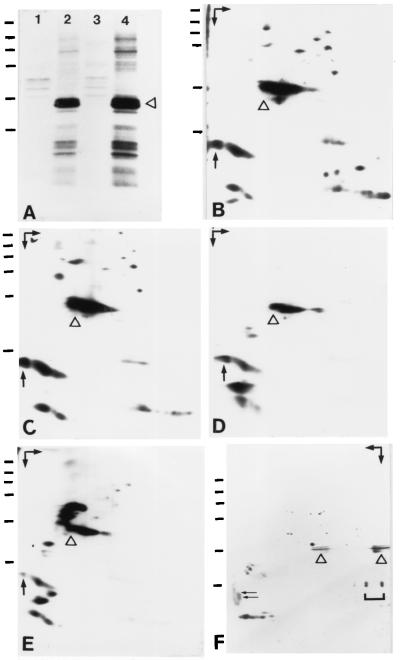
l-[35S]Methionine-labeled A6 cells were lysed in buffer containing 250 mM NaCl and separated by centrifugation into supernatant and pellet fractions. Under these conditions, protein NO38 was almost completely solubilized, as determined by immunoblotting of the resulting fractions. The supernatant was then subjected to immunoprecipitation using mAbs to protein NO38 or control antibodies of the same IgG subclass, all bound to protein G-Sepharose, and immunoprecipitates were analyzed by one- and 2D gel electrophoresis and autoradiography. Certain low molecular weight proteins were specifically coprecipitated with NO38, whereas some high molecular weight proteins detectable in the immunoprecipitates were bound unspecifically to protein G-Sepharose (Fig. 1A). 2D gel electrophoresis revealed that the low molecular weight proteins consist of acidic as well as basic polypeptides (Fig. 1B). By comigration of l-[35S]methionine-labeled immunoprecipitates with unlabeled proteins from 40S and 60S subunits of mature ribosomes on 2D-PAGE, the basic proteins could clearly be identified as r-proteins (data not shown). Under more stringent buffer conditions (400 mM NaCl), the r-proteins were removed partially from the immunocomplexes (Fig. 1C). Further increase of stringency by including a wash in high salt buffer prior to electrophoresis of the immunoprecipitates resulted in quantitative depletion of the basic proteins (Fig. 1D). Surprisingly, the newly detected acidic proteins remained bound to the protein NO38 immunocomplexes under either condition, indicating that they might represent a complex of tightly associated proteins.
Figure 1.
Coimmunoprecipitations and affinity chromatography using mAbs (No-63 and No-185) against nucleolar protein NO38 of X. laevis. (A) Autoradiograph of immunoprecipitates obtained with mAb No-63 (lane 2) or mAb No-185 (lane 4) from extracts of [35S]methionine-labeled A6 cells, prepared in lysis buffer containing 250 mM NaCl and separated by SDS/PAGE. Both mAbs coprecipitated, besides protein NO38 (denoted by a triangle), a group of low molecular weight proteins (Mr ≤ 29,000). By contrast, proteins of higher molecular weight (Mr ≥ 45,000) present in such immunoprecipitates were bound unspecifically to protein G-Sepharose (lanes 1 and 3). (B–E) Autoradiographs of immunoprecipitates obtained with mAb No-63 from cellular extracts (B–D) and extracted nuclear LSP (E) of [35S]methionine-labeled A6 cells, analyzed by 2D gel electrophoresis (first dimension: NEPHGE). The position of nucleolar protein NO38 is indicated by a triangle. (B) Analysis of the immunoprecipitates shown in A (lane 2) disclosed the acidic as well as basic nature of the low molecular weight proteins coprecipitated with protein NO38. (C and D) Basic proteins were removed partially at higher ionic strength (400 mM NaCl; C) and completely by treatment of the immunoprecipitates with high-salt buffer (D, PBS plus 1 M NaCl). By contrast, the acidic proteins remained in the immunoprecipitates. (E) After immunoprecipitation of proteins extracted from LSP fractions of isolated A6 nuclei with buffer containing 400 mM NaCl, only the acidic proteins coprecipitated whereas basic proteins were not detectable. (F) Coomassie blue-stained proteins obtained by preparative affinity chromatography (mAb No-185) from unlabeled cellular extract, prepared as in A, after 2D gel electrophoresis (first dimension: IEF). The position of nucleolar protein NO38 is indicated by a triangle. The bracket marks IgG light chains leached from the No-185 antibody column. The largest and most acidic of the co-precipitating proteins, denoted by an arrow in B–E, was resolved into two closely migrating polypeptides of Mr 24,000 and 23,000 (pI ≈ 4.3), marked by two arrows in F. Arrows in the upper corners in B–F indicate the directions of electrophoresis (first dimension, NEPHGE or IEF, horizontal arrow; second dimension, SDS/PAGE, vertical arrow). Basic proteins are in the right-hand part of each protein separation. Horizontal bars indicate reference proteins of 205, 116, 97.4, 66, 45, and 29 kDa (from top to bottom).
To examine the existence of such a complex, we repeated the immunoprecipitations using extracts obtained from LSPs of A6 nuclear homogenates under stringent conditions. Here, only the acidic coprecipitating proteins mentioned were found to cosediment with protein NO38 (Fig. 1E), whereas basic proteins were completely absent. The same result was obtained when nuclear extracts were fractionated by sucrose gradient centrifugation or gel filtration chromatography, and the recovered 7S or Mapp 600,000 fractions known to contain protein NO38 (14, 33) were used for immunoprecipitation.
Isolation and cDNA Cloning of an Acidic Protein Coprecipitating with Protein NO38.
To elucidate the nature of these proteins, we isolated protein NO38 and associated proteins in a preparative scale from unlabeled extracts of approximately 4 × 108 A6 cells by affinity chromatography using mAb No-185 covalently coupled to Sepharose beads. Because we were most interested in the identification of the acidic proteins, the immunoisolated protein complexes were separated by 2D-PAGE and transferred to PVDF membranes (Fig. 1F). After staining with Coomassie blue, the two most acidic polypeptides (Mr 24,000 and 23,000, both of pI ≈ 4.3) were individually subjected to amino acid sequencing. Two amino acid sequences, EVTVPLANLK and DEHNVVEVTAPN, were obtained from each polypeptide, indicating that they may represent two different isoforms—or degradation products—of the same protein. Database searches for both oligopeptides were negative. Using degenerated antisense primers deduced from the peptide sequence DEHNVVEVTAPN and the library-specific reverse primer as sense primer, we PCR-amplified a 249-nt cDNA fragment from a cDNA library from X. laevis kidney. This cDNA fragment was then used as a probe for screening the same cDNA library. Sixteen positive clones contained identical cDNA inserts of ≈1.6 kb, as judged from their electrophoretic mobilities and from partial sequencing analyses; one of them, termed pBT-NO29, was used for further analyses.
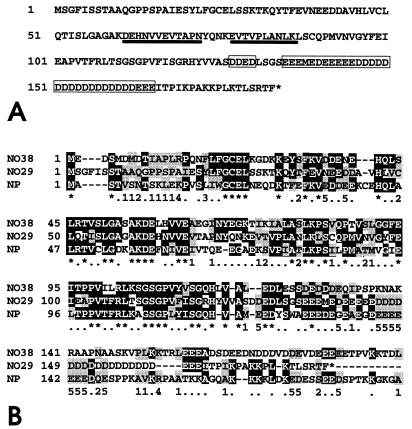
The nucleotide sequence has been determined on both strands and the aa sequence deduced therefrom is shown in Fig. 2A. The clone consists of 1,593 nt, contains a 5′-untranslated region of 63 nt meeting the −3 = A requirement of Kozak (44), an ORF of 549 nt (nt 64–612), and a 3′ region of 981 untranslated nt (nt 613-1,593) with a poly(A)+ addition signal 18 bp upstream of the poly(A) tail, of which 26 nt (nt 1,568–1,593) are included. The ORF encodes a polypeptide of 183 aa with a predicted Mr of 20,121 containing the two oligopeptides, EVTVPLANLK and DEHNVVEVTAPN (Fig. 2A), initially obtained by amino acid sequencing. The most conspicuous feature of protein NO29 is the presence of a large cluster of 30 negatively charged amino acid residues near the C-terminus (positions 135–165), interrupted only by the single methionine at position 138. Together with 17 additional glutamic and aspartic acid residues scattered throughout the entire sequence, the protein contains 47 acidic amino acids, comprising 25.6% of the total protein and contributing to the predicted very low pI of 3.75.
Figure 2.
Amino acid sequence of protein NO29 and comparison with its homologs, protein NO38 and nucleoplasmin. (A) Amino acid sequence (single-letter code) of protein NO29 of X. laevis deduced from the cDNA insert of pBT-NO29 (accession no. Z85983; nt 64–612). The protein consists of 183 aa, including the N-terminal methionine. The acidic cluster in the C-terminal part of the molecule is indicated by boxes, and the two peptide sequences identified by amino acid sequencing are underlined. (B) Sequence comparison between the X. laevis proteins NO29, NO38, and nucleoplasmin. Identical and conservatively exchanged amino acids are shaded black and grey, respectively. Horizontal bars are omissions introduced to optimize the alignment. The symbols line below the shaded sequences are defined as follows: ∗, identical amino acids in all three proteins; ⋅, identical amino acids in two of the proteins; numbers 1–5 define conservative exchanges (1, aliphatic, nonpolar; 2, aliphatic, polar; 3, aromatic; 4, basic; 5, acidic).
Database searches disclosed a striking homology of protein NO29 to NO38 and nucleoplasmin (for sequence alignment, see Fig. 2B): protein NO38 shares 65 identical and 27 conservatively exchanged amino acids with protein NO29 throughout the entire protein, resulting in 50% homology. Nucleoplasmin shares 65 identical and 35 conservatively exchanged amino acids, resulting in 54% homology. These findings indicated that we had discovered another member of the nucleoplasmin family.
Molecular and Biochemical Characterization of Protein NO29.
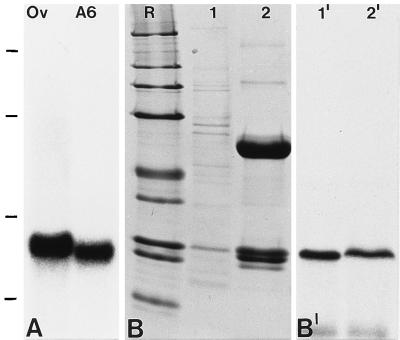
Northern blot hybridization on poly(A)+ RNA from X. laevis ovaries and A6 cells yielded a strong signal at ≈1.7 kb (Fig. 3A), indicative of a full-length cDNA clone. The authenticity of the 5′ end of the isolated cDNA was further verified by two independent 5′-RACE (rapid isolation of cDNA ends) experiments using poly(A)+ mRNA isolated from A6 cells as template (data not shown). In vitro transcription/translation of pBT-NO29 mRNA resulted in a distinct translation product with Mr 29,000 on SDS/PAGE (Fig. 3 B and B′, lanes 1 and 1′), clearly different from the predicted Mr of 20,121. Such obvious deviations of Mr estimations and predicted Mr values have also been reported for several proteins containing clusters of negatively charged aa (nucleoplasmin, proteins N1/N2 and NO38, nucleolin) and appear to be due to lesser SDS binding and hence an anomalously low electrophoretic mobility (14, 20, 45). The protein synthesized in vitro could be efficiently immunoprecipitated with NO29 antibodies (Fig. 3 B and B′,; lanes 2 and 2′), thus confirming both the identity of the translation product and the specificity of the antibodies. On 2D-PAGE, the in vitro synthesized, [35S]methionine-labeled protein NO29 comigrated with a single polypeptide of Mr 29,000 and pI ≈ 4.1 present in A6 cells, as detected by immunoblotting (Fig. 4 A–A"). Because the two additonal immunoreactive polypeptides of Mr 24,000 and 23,000, both of pI ≈ 4.3 (Fig. 4A′) migrate at the same position as the most acidic polypeptides of the immunocomplexes (Fig. 1F), they probably represent degradation products of protein NO29.
Figure 3.
Molecular characterization of the mRNA and polypeptide encoded by cDNA clone pBT-NO29. (A) Autoradiograph showing a Northern blot hybridization with an antisense riboprobe for protein NO29 on poly(A)+ RNA from X. laevis ovary (Ov) or A6 cells (A6). Note the specific reaction with an ≈1.7-kb RNA. RNA size markers of 7.5, 4.4, 2.4, and 1.4 kb are indicated on the left margin (from top to bottom). (B) Coomassie blue staining of SDS/PAGE-separated proteins from a rabbit reticulocyte lysate used for in vitro transcription/translation of clone pBT-NO29 (lane 1) and of the immunoprecipitate from the same translation mixture using antibodies to protein NO29 (lane 2; the prominent polypeptides represent heavy and light Ig chains). Lane designated R shows reference proteins of 205, 116, 97.4, 66, 45, 36, 29, 24, and 20 kDa (from top to bottom). (B′) Corresponding autoradiograph showing the translation product of Mr ≈ 29,000 (lane 1′), which can be immunoprecipitated by antibodies to protein NO29 (lane 2′).
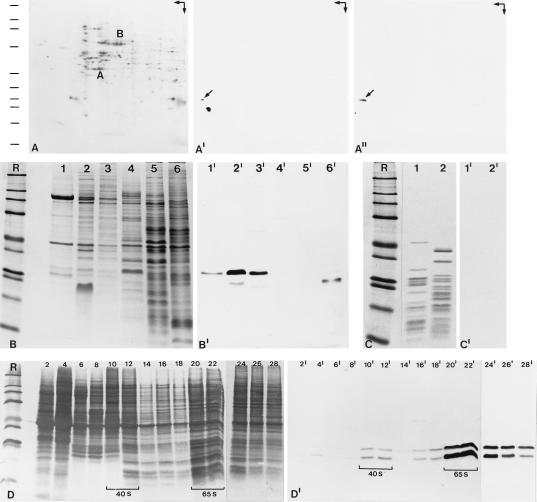
Figure 4.
Biochemical characterization of protein NO29. (A–A") 2D gel electrophoresis (arrows in the upper right corner indicate directions of first dimension, IEF, horizontal, and second dimension, SDS/PAGE, vertical) of total proteins from A6 cells in coelectrophoresis with the [35S]methionine-labeled protein NO29 obtained by translation in vitro, followed by transfer of the separated proteins to a PVDF membrane and detection by Coomassie blue staining (A), immunoblotting with protein NO29 antibodies (A′), or autoradiography (A"). The positions of α-actin (A) and BSA (B) are indicated. The antibodies react with a very acidic polypeptide of Mr 29,000 (A′, arrow) which comigrates with protein NO29 encoded by pBT-NO29 (A", arrow). The two additional immunoreactive polypeptides of Mr 24,000 and 23,000 most likely represent degradation products of protein NO29. Reference proteins in A–D, indicated by bars in A, are the same as in Fig. 3B (lanes designated R). (B–B") Identification of protein NO29 in cellular and nuclear fractions from oocytes and somatic cells of X. laevis. (B) Coomassie blue staining of SDS/PAGE-separated proteins from total oocyte nuclei (lane 1); proteins of LSP, HSP, and HSS fractions from an oocyte nuclear homogenate (lanes 2–4); egg extract proteins (lane 5); and total proteins from A6 cells (lane 6). (B′) Corresponding immunoblot with antibodies to protein NO29 react specifically with a polypeptide of Mr 29,000 present among the nuclear proteins, notably the LSP and HSP fractions (lanes 1′–3′), which is not detectable in the HSS fraction (lane 4′). In extracts from eggs and A6 cells (lanes 5′ and 6′), this reaction is detected only after longer exposure, partly (lane 6′) in a degradation product. (C) Coomassie blue-stained proteins after SDS/PAGE of components of 40S (lane 1) and 60S (lane 2) subunits of cytoplasmic ribosomes from Xenopus ovary. (C′) Corresponding immunoblot with antibodies to protein NO29 reveals its absence from mature ribosomes. (D and D′) Identification of protein NO29 in free nucleoplasmic 40S and 65S pre-ribosomal particles, separated by sucrose gradient centrifugation of HSP fractions from Xenopus oocyte nuclei. (D) Coomassie blue staining of the resulting gradient fractions (fraction numbers are indicated). (D′) Corresponding immunoblot, showing that protein NO29 is recovered in both 40S and 65S preribosomal particles (denoted by brackets).
The distribution of protein NO29 in different fractions from Xenopus oocyte nuclei, egg extracts, and somatic cells was analyzed by immunoblotting with NO29-specific antibodies. The protein was detected in total nuclei, in LSP and HSP fractions (Fig. 4 B and B′, lanes 1, 1′, 2, 2′, 3, and 3′), indicating that the protein is a nuclear protein bound to a relatively large structure. In contrast, the protein was not detected in the HSS fraction and present only in trace amounts in egg extracts (Fig. 4 B and B′, lanes 4, 4′, 5, and 5′). As the protein NO29-containing HSP fractions contain preribosomes, we fractionated them further on 10–40% sucrose gradients and analyzed the resulting fractions for the protein by SDS/PAGE and immunoblotting (Fig. 4 D and D′). Both the 29-kDa protein and the two major degradation products were specifically enriched in fractions containing the precursors for the small and the large ribosomal subunits (“preribosomal particles”), recognized as 40S and 65S particles (Fig. 4 D and D′). On the other hand, protein NO29 was not present in detectable amounts in mature cytoplasmic ribosomes, as determined by immunoblotting experiments on 40S and 60S ribosomal subunits (Fig. 4 C and C′) and by immunocytochemistry.
Intracellular Localization of NO29.
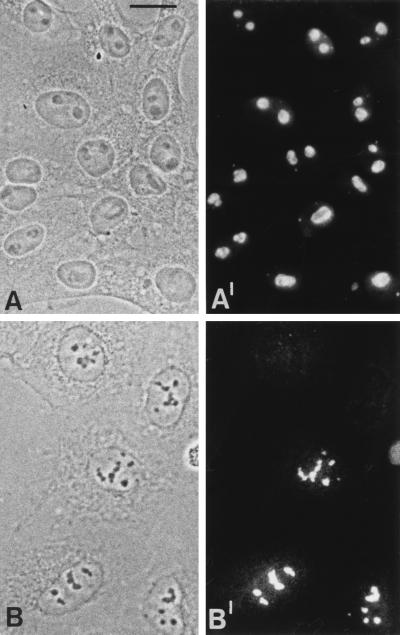
Immunofluorescence microscopy of A6 cell cultures using antibodies to protein NO29 showed the protein specifically accumulated in nucleoli, with some concentration in the granular portion, and to a lesser extent, in some cells, finely dispersed over the nucleoplasm (Fig. 5 A and A′). Moreover, protein NO29 was detected as a constitutive component in the nucleoli of diverse X. laevis cell types, from intestinal epithelium to oocytes, and also in the residual nucleoli of erythrocytes (data not shown). This result was confirmed by transient and stable transfection of human carcinoma cells of line PLC with a cDNA encoding an aminoterminally myc-tagged protein NO29. Immunofluorescence analysis using myc antibodies revealed that the ectopically expressed protein was exclusively located in the nucleoli of the transfected cells (Fig. 5 B and B′).
Figure 5.
Immunolocalization of protein NO29. Phase contrast (A and B) and corresponding immunofluorescence (A′ and B′) micrographs are shown. Cultured X. laevis A6 cells reacted with protein NO29 antibodies (A and A′) and human PLC-cells transfected with a cDNA construct encoding myc-tagged protein NO29 (B and B′), detected by myc-specific mAb 9E10. Protein NO29 is markedly accumulated in nucleoli of A6 cells. Only in some cells an additional faint nucleoplasmic staining was detected. In transfected human PLC cells, the Xenopus protein NO29 localizes exclusively to nucleoli. (Bar in A = 20 μm. Identical magnifications in all pictures.)
Upon treatment of A6 cells with actinomycin D, protein NO29 appeared to be partly released from the nucleoli, first to the nucleoplasm and then to the cytoplasm (data not shown), as it has also been reported for protein NO38/B23 after treatment with this and other cytotoxic drugs (46, 47).
DISCUSSION
The nucleolar protein NO29 described here is a member of what seems to represent the nucleoplasmin family of proteins, its amino acid sequence being almost equally distant from both the diffusible karyoplasmic protein, nucleoplasmin, and the constitutive nucleolar protein, NO38/B23. Specifically, however, protein NO29 is characterized by its small size of only 183 aa, compared with 200 aa in nucleoplasmin and 299 aa in protein NO38, and by its exceptionally low isoelectric point of 3.75, making it the most acidic nuclear protein known. Its high negative charge, in particular the large cluster of glutamic and aspartic acid residues (34 of 39 aa) in the segment preceeding the 18-aa C-terminal stretch, and its location in the nucleolus and in preribosomal particles, suggest that protein NO29 may bind to basic proteins such as r-proteins as nucleoplasmin binds histones H2A and H2B (ref. 5; for protein NO38/B23 in preribosomes see refs. 14, 19, and 21). However, although we could demonstrate mixed complexes containing both proteins NO29 and NO38, we have yet to identify direct complexes of protein NO29 with any r-protein or with nucleic acids (for binding of protein NO38/B23 to DNA and RNA, see refs. 23, 25, and 26). In discussing possible binding partners of protein NO29, it should further be considered that the related protein NO38/B23 has been reported to bind to various other nucleolar proteins such as protein p120 (31) or to the nonnucleolar transcription factor YY1 (27) as well as to certain viral proteins such as Rev and Tat of HIV-1 or the human T-lymphocyte virus type I protein Rex (28–30, 48–50).
Moreover, the detection of protein NO29 in the nucleoli of many diverse cell types, from oocytes to cultured cells and from intestinal epithelial cells to the residual nucleoli of erythrocytes, indicates that this protein is a constitutive and functionally important nucleolar component, although its relative abundance is more than an order of magnitude below that of protein NO38/B23. In computer-based expressed sequence tag searches, we have also noticed the existence of a mouse mRNA encoding another protein of this family, showing significant amino acid sequence homology to both protein NO29 and NO38, that obviously is related to the Npm3 gene product recently described (51). We are currently attempting to identify the mammalian cDNA clones orthologous to amphibian NO29 and to examine possible functions of protein NO29 by genetic approaches.
In view of the general sequence similarity of the three nuclear proteins, nucleoplasmin, NO38/B23 and NO29 (Fig. 2), it is remarkable to note that protein NO29 does not display significant homology to the nuclear localization signal (NLS)-containing aa sequences of the other two molecules. Whether protein NO29 contains a functioning NLS (a candidate hexapeptide KPAKKP is seen in positions 170–174) or whether it is transported into the nucleolus by binding to NLS carrying other proteins (for a review, see ref. 13) will be examined by targeted mutagenesis.
Similarly surprising in protein NO29 is the absence of the entire C-terminal portion of protein NO38/B23 which has been reported to contribute essentially to the accumulation of this protein in nucleoli (32, 33) and to its nucleic acid binding (25). On the other hand, our finding (results not shown) that protein NO29 does not form the distinct oligomers described for protein NO38/B23 (14, 21) and nucleoplasmin (20) shows that the N-terminal portion, despite the considerable sequence homology between these proteins, including the region between amino acid positions 15–120 and in particular the cysteine residue at position 22 of protein NO38 (for details, see ref. 33) is essentially different in protein NO29. Hence, the specific topogenically and functionally important element in protein NO29 cannot be hypothesized from sequence similarities but will have to be determined experimentally.
Acknowledgments
We thank Sylvia Knecht for expert technical assistance, Hans Heid and Martina Schnölzer for superior protein sequencing and mass spectrometry, Hans Richard Rackwitz for preparing and keyhole limpet hemocyanin coupling of synthetic peptides, Jutta Müller-Osterholt for preparing the photographs and Eva Ouis for careful typing of the manuscript. This study has been supported by the Deutsche Forschungsgemeinschaft (Grant Schm 862/2-3 to M.S.S.-Z.).
ABBREVIATIONS
- r-proteins
ribosomal proteins
- LSP
low speed pellet
- HSP
high speed pellet
- HSS
high speed supernatant
- IEF
isoelectric focusing
- NEPHGE
nonequilibrium pH gradient electrophoresis
- RT
room temperature
- 2D
two dimensional
- PVDF
polyvinylidene difluoride
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Z85983).
References
- 1.Laskey R A, Honda B M, Mills A D, Finch J T. Nature (London) 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 2.Krohne G, Franke W W. Proc Natl Acad Sci USA. 1980;77:1034–1038. doi: 10.1073/pnas.77.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills A D, Laskey R A, Black P, De Robertis E M. J Mol Biol. 1980;139:561–568. doi: 10.1016/0022-2836(80)90148-5. [DOI] [PubMed] [Google Scholar]
- 4.Kleinschmidt J A, Franke W W. Cell. 1982;29:799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- 5.Kleinschmidt J A, Fortkamp E, Krohne G, Zentgraf H, Franke W W. J Biol Chem. 1985;260:1166–1176. [PubMed] [Google Scholar]
- 6.Cotten M, Chalkley R. EMBO J. 1987;6:3945–3954. doi: 10.1002/j.1460-2075.1987.tb02736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith S, Stillman B. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 8.Ishimi Y, Hirosumi J, Sato W, Sugasawa K, Yokota S, Hanaoka F, Yamada M. Eur J Biochem. 1984;142:431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- 9.Sapp M, Worcel A. J Biol Chem. 1990;265:3957–9365. [PubMed] [Google Scholar]
- 10.Gruss C, Sogo J M. BioEssays. 1992;14:1–8. doi: 10.1002/bies.950140102. [DOI] [PubMed] [Google Scholar]
- 11.Scheer U, Benavente R. BioEssays. 1990;12:14–21. doi: 10.1002/bies.950120104. [DOI] [PubMed] [Google Scholar]
- 12.Olson M. In: The Eucaryotic Nucleus. Strouss P R, Wilson S H, editors; Strouss P R, Wilson S H, editors. Vol. 2. Caldwell, NY: Telford; 1990. pp. 519–559. [Google Scholar]
- 13.Xue Z, Mélèse T. Trends Cell Biol. 1994;4:414–417. doi: 10.1016/0962-8924(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt-Zachmann M S, Hügle-Dörr B, Franke W W. EMBO J. 1987;6:1881–1890. doi: 10.1002/j.1460-2075.1987.tb02447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busch H. In: Chromosomal Nonhistone Proteins, Vol. IV, Structural Associations. Hnilica L S, editor; Hnilica L S, editor. Boca Raton, FL: CRC; 1984. pp. 233–286. [Google Scholar]
- 16.Chan P-K, Chan W-Y, Yung B Y M, Cook R G, Aldrich M B, Ku D, Goldknopf I L, Busch H. J Biol Chem. 1986;261:14335–14341. [PubMed] [Google Scholar]
- 17.Chang J-H, Dumbar T S, Olson M O J. J Biol Chem. 1988;263:12824–12827. [PubMed] [Google Scholar]
- 18.Schmidt-Zachmann M S, Franke W W. Chromosoma. 1988;96:417–426. doi: 10.1007/BF00303035. [DOI] [PubMed] [Google Scholar]
- 19.Chan P-K, Liu Q R, Durban E. Biochem J. 1990;270:549–552. doi: 10.1042/bj2700549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingwall C, Dilworth S M, Black S J, Kearsey SE, Cox L S, Laskey R A. EMBO J. 1987;6:69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yung B J-M, Chan P-K. Biochim Biophys Acta. 1987;925:74–82. doi: 10.1016/0304-4165(87)90149-8. [DOI] [PubMed] [Google Scholar]
- 22.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 23.Dumbar T S, Gentry G A, Olson M O J. Biochemistry. 1989;28:9495–9501. doi: 10.1021/bi00450a037. [DOI] [PubMed] [Google Scholar]
- 24.Feuerstein N, Mond J J, Kinchington P R, Hickey R, Karjalainen Lindsberg M-L, Hay I, Ruyechan W T. Biochim Biophys Acta. 1990;1087:127–136. doi: 10.1016/0167-4781(90)90196-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Baumann A, Szebeni A, Olson M O J. J Biol Chem. 1994;269:30994–30998. [PubMed] [Google Scholar]
- 26.Herrera J E, Savkur R, Olson M O J. Nucleic Acids Res. 1995;23:3974–3979. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inouye C J, Seto E. J Biol Chem. 1994;269:6506–6510. [PubMed] [Google Scholar]
- 28.Fankhauser C, Izaurralde E, Adachi Y, Wingfield P, Laemmli U K. Mol Cell Biol. 1991;11:2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dundr M, Leno G H, Hammarskjöld M-L, Rekosh D, Helga-Maria C, Olson M O J. J Cell Sci. 1995;108:2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- 30.Miyazaki Y, Takamatsu T, Nosaka T, Fujita S, Martin T E, Hatanaka M. Exp Cell Res. 1995;219:93–101. doi: 10.1006/excr.1995.1209. [DOI] [PubMed] [Google Scholar]
- 31.Valdez B C, Perlaky L, Henning D, Saijo Y, Chan P-K, Busch H. J Biol Chem. 1994;269:23776–23783. [PubMed] [Google Scholar]
- 32.Peculis B A, Gall J G. J Cell Biol. 1992;116:1–14. doi: 10.1083/jcb.116.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zirwes R F, Kouzmenko A P, Peters J-M, Franke W W, Schmidt-Zachmann M S. Mol Biol Cell. 1997;8:231–248. doi: 10.1091/mbc.8.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Köhler A, Schmidt-Zachmann M S, Franke W W. J Cell Sci. 1997;110:1051–1062. doi: 10.1242/jcs.110.9.1051. [DOI] [PubMed] [Google Scholar]
- 35.Hügle B, Scheer U, Franke W W. Cell. 1985;41:615–627. doi: 10.1016/s0092-8674(85)80034-9. [DOI] [PubMed] [Google Scholar]
- 36.Evans J P, Kay B K. Methods Cell Biol. 1991;36:138–139. doi: 10.1016/s0091-679x(08)60275-7. [DOI] [PubMed] [Google Scholar]
- 37.Cordes V C, Reidenbach S, Köhler A, Stuurman N, van Driel R, Franke W W. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hügle B, Hazan R, Scheer U, Franke W W. J Cell Biol. 1985;100:873–886. doi: 10.1083/jcb.100.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miake-Lye R, Kirschner M W. Cell. 1985;41:165–175. doi: 10.1016/0092-8674(85)90071-6. [DOI] [PubMed] [Google Scholar]
- 40.Mertens C, Kuhn C, Franke W W. J Cell Biol. 1996;135:1009–1025. doi: 10.1083/jcb.135.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 42.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt-Zachmann M S, Nigg E A. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- 44.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 45.Kleinschmidt J A, Dingwall C, Maier G, Franke W W. EMBO J. 1986;5:3547–3552. doi: 10.1002/j.1460-2075.1986.tb04681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yung B Y-M, Busch R K, Busch H, Mauger A B, Chan P-K. Biochem Pharmacol. 1985;34:4059–4063. doi: 10.1016/0006-2952(85)90387-9. [DOI] [PubMed] [Google Scholar]
- 47.Chan P-K. Exp Cell Res. 1992;203:174–181. doi: 10.1016/0014-4827(92)90053-b. [DOI] [PubMed] [Google Scholar]
- 48.Cochrane A W, Perkins A, Rosen C A. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siomi H, Shida H, Maki M, Hatanaka M. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adachi Y, Copeland T D, Hatanaka M, Oroszlan S. J Biol Chem. 1993;268:13930–13934. [PubMed] [Google Scholar]
- 51.MacArthur C A, Shackleford G M. Genomics. 1997;42:137–140. doi: 10.1006/geno.1997.4353. [DOI] [PubMed] [Google Scholar]