Abstract
A novel type I transmembrane protein of COPI-coated vesicles, p23, has been demonstrated to be localized mainly to the Golgi complex. This protein and p24, another member of the p24 family, have been shown to bind coatomer via their short cytoplasmic tails. Here we demonstrate that p23 continuously cycles through the early secretory pathway. The cytoplasmic tail of p23 is shown to act as a functional retrieval signal as it confers endoplasmic reticulum (ER) residence to a CD8–p23 fusion protein. This ER localization is, at least in part, a result of retrieval from post-ER compartments because CD8–p23 fusion proteins receive post-ER modifications. In contrast, the cytoplasmic tail of p24 has been shown not to retrieve a CD8–p24 fusion protein. The coatomer binding motifs FF and KK in the cytoplasmic tail of p23 are reported to influence the steady-state localization of the CD8–p23 fusion protein within the ER–Golgi recycling pathway. It appears that the steady-state Golgi localization of endogenous p23 is maintained by its lumenal domain, as a fusion protein with the lumenal domain of CD8, and the membrane span as well as the cytoplasmic tail of p23 is no longer detected in the Golgi.
Keywords: membrane traffic, vesicular transport, protein retrieval, retrograde transport, coated vesicles
Biosynthetic protein transport along the secretory pathway is mediated by vesicular carriers that bud off from a donor compartment and fuse with an acceptor compartment thereby delivering secretory cargo molecules (1). The formation of these transport vesicles is mediated by the recruitment of specific coat proteins that may act as a mechanical device to form a spherical vesicle from a flattened donor membrane (2). Two types of coat structures have been characterized that are involved in biosynthetic protein transport. COPI was described to mediate anterograde endoplasmic reticulum (ER) to Golgi transport (3–5), intra-Golgi transport (6), as well as retrograde Golgi to ER transport (7–10). COPII appears to mediate exclusively anterograde ER to Golgi transport (11).
A novel family of type I transmembrane proteins, the p24 family, has been defined by the COPI vesicle protein chop24 (12) and a putative yeast homologue Emp24p (13). Database searches revealed the existence of a number of relatives in both yeast and mammals with at least five homologues within one species. All members are composed of a large lumenal domain, a single membrane stretch, and a short cytoplasmic tail. The steady-state localization of only a few p24 proteins is known: p23 is localized to the Golgi complex and, to a lower extent, the intermediate compartment (IC) (14), and some yeast p24 proteins are predominantly localized to the ER (13, 15). The cytoplasmic tails of many p24 proteins contain one or two phenylalanine residues close to the membrane, whereas only a subset of family members contains a double lysine motif close to the C-terminus (14, 16). The cytoplasmic tails of many p24 proteins were shown to bind coatomer, the cytosolic precursor of the COPI-coat, similar to the KKXX retrieval motifs present in the cytoplasmic tails of ER resident type I membrane proteins. However, the characteristics of coatomer binding are different, as coatomer binding to p24 tails depends on an FF motif and is modulated by a KK motif (14, 16), whereas the binding of coatomer to ER residents depends only on their double lysine motif (7, 17).
Functional studies on p24 proteins suggest a role in the budding process of transport vesicles (12, 13, 18). Chop24 was described to be a component of COPI-coated vesicles isolated from CHO cells. When a putative yeast homologue of chop24, Emp24p, was deleted the corresponding yeast strain was still viable. However, in combination with a temperature-sensitive allele of the N-ethylmaleimide-sensitive fusion protein (NSF), deletion of Emp24p resulted in a decreased accumulation of transport vesicles at the restrictive temperature, indicating that disruption of the chop24 homologue affects the formation of transport vesicles (12). Another study on Emp24p indicates that the protein is also a component of COPII-coated vesicles and a deletion of the corresponding gene caused a transport block of a subset of secretory proteins (13). In addition, p24 proteins were cloned as suppressors of mutations in the COPII subunit SEC13 and proposed to regulate COPII vesicle formation (18). Thus, p24 proteins are discussed to function in the selection of cargo proteins dependent on the proper assembly of COPII-coated vesicles. Finally, a putative yeast homologue of p23, Erv25p, has been described to physically interact with Emp24p. This interaction was proposed to be important for efficient anterograde ER to Golgi transport (15). In general, p24 proteins could serve, in addition to coat binding, as cargo receptors that are needed to efficiently deliver proteins along the secretory pathway.
In the present study the intracellular dynamics of p23 are investigated. Although p23 is mainly localized to the Golgi at steady-state, we demonstrate that the protein continuously cycles through the early secretory pathway. Accordingly, we have identified a functional retrieval signal in the cytoplasmic tail of p23 that depends on its two coatomer binding motifs.
MATERIALS AND METHODS
Antibodies.
Affinity-purified antibodies directed against the C terminus of p23 (no. 1402) were used as described (14). A polyclonal antiserum against rat liver Golgi membranes was generated in the following way: Golgi membranes were enriched according to (19, 20) by a two-step purification protocol using glucosylceramide synthase activity as Golgi marker. After high salt- and carbonate treatment membrane proteins were prepared by phase partitioning in Triton X-114 and used to immunize rabbits according to standard procedures. The obtained antiserum was incubated with purified ER membranes to remove ER-specific antibodies. In immunofluorescence experiments, the corresponding supernatant specifically stains the Golgi complex as judged by colocalization with Golgi-bound coatomer [CM1 (ref. 14, and data not shown). Monoclonal antibodies directed against the lumenal domain of CD8 were prepared from the cell culture supernatant of the cell line OKT 8 (ATCC CRL-8014). Polyclonal rabbit antibodies directed against the ER marker calreticulin and monoclonal mouse antibodies directed against the IC marker ERGIC53 were the generous gifts of Hans-Dieter Söling (University of Göttingen, Göttingen, Germany) and Hans-Peter Hauri (Biocenter Basel, Basel), respectively. Polyclonal rabbit α-lamp-1 antibodies (used to detect lysosomes) were kindly provided by Walter Hunziker (University of Lausanne, Lausanne, Switzerland) and originated from Ira Mellman (Yale University).
Construction of CD8–p23 Fusion Proteins.
All cloning procedures were performed according to standard procedures. The eukaryotic expression vector pCMUIV containing the CD8 cDNA (21) was used for all constructs. The DNA sequence encoding the membrane-spanning domain as well as the cytoplasmic tail of CD8 was replaced by synthetic oligonucleotides encoding the corresponding sequences of p23 (ApaLI–BamHI fragment). Alternatively, only the DNA sequence encoding the cytoplasmic tail of CD8 was replaced by the corresponding sequence of p23 (SalI–BamHI fragment). Mutations in the coatomer binding motifs were introduced by the use of synthetic oligonucleotides encoding the corresponding amino acid changes. The predicted nucleotide sequence of all constructs was confirmed by sequencing using the dideoxynucleotide method.
Cell Culture, Transfection Procedures, and Indirect Immunofluorescence.
NRK, COS, and HeLa cells were grown according to standard conditions. For transient expression of cDNAs cells were grown on glass coverslips. DNA transfer was performed using the calcium phosphate precipitation method according to Nilsson et al. (21). Further processing was performed 24 h posttransfection. Where indicated cells were incubated at 15°C (using Hepes-buffered medium) to inhibit anterograde IC to Golgi transport (22). Cells were then prepared for indirect immunofluorescence according to standard protocols including methanol fixation/permeabilization and paraformaldehyde fixation/Triton X-100 permeabilization. After incubation of primary (see above) and secondary antibodies (Dianova, Hamburg, Germany) cells were washed and embedded in Fluoromount G (Biozol, Eching, Germany). Samples were viewed using a Zeiss Axiovert 35 microscope equipped with the appropriate filters for fluorescein isothiocyanate- and tetramethylrhodamine B isothiocyanate-derived fluorescence.
Pulse–chase Analysis of CD8–p23 Fusion Proteins.
Pulse–chase analysis was performed according to Jackson et al. (23). Briefly, COS cells were grown on culture dishes and transfected with the various CD8–p23–p23 fusion proteins using the calcium phosphate precipitation method. Twenty-four hours posttransfection, cells were labeled with 150 μCi/ml (1 Ci = 37 GBq) [35S]methionine/cysteine (Amersham) for 30 min. The cells were then either kept on ice or further incubated at 37°C for 30 min in chase medium containing methionine/cysteine at a final concentration of 10 mM. Cell were lysed in buffer containing 1% TX-100. After removing unsoluble material, CD8–p23 fusion proteins were immunoprecipitated using monoclonal antibodies directed against the lumenal domain of CD8 (OKT8). After separation on 12% SDS/polyacrylamide gels (12 × 15 cm) precipitates were analyzed by autoradiography using β-max hyperfilms (Amersham).
RESULTS
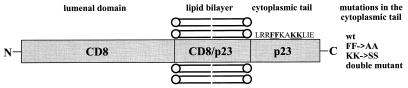
To characterize targeting signals in the cytoplasmic tail of p23 we have constructed fusion proteins composed of (i) the lumenal domain of the cell surface protein CD8 (24) and the membrane-spanning domain as well as the cytoplasmic tail of p23, and ii) the lumenal plus transmembrane domains of CD8 and the cytoplasmic tail of p23. Furthermore, the coatomer binding motifs in the cytoplasmic tail of p23 (14) were mutated. The various versions of CD8–p23 fusion proteins (Fig. 1) were compared with similar CD8–p24 fusion proteins (16).
Figure 1.
Schematic description of the various versions of CD8–p23 fusion proteins. The constructs are divided into two classes either containing the CD8 (referred to as CD8–CD8–p23) or the p23 (referred to as CD8–p23–p23) membrane-spanning domain. The cytoplasmic tail of p23 was introduced into four different versions: (i) wild-type sequence, (ii) FF to AA mutation, (iii) KK to SS mutation, and (iv) double mutation (both FF to AA and KK to SS mutation).
The Cytoplasmic Tail of p23 Contains a Functional Retrieval Signal.
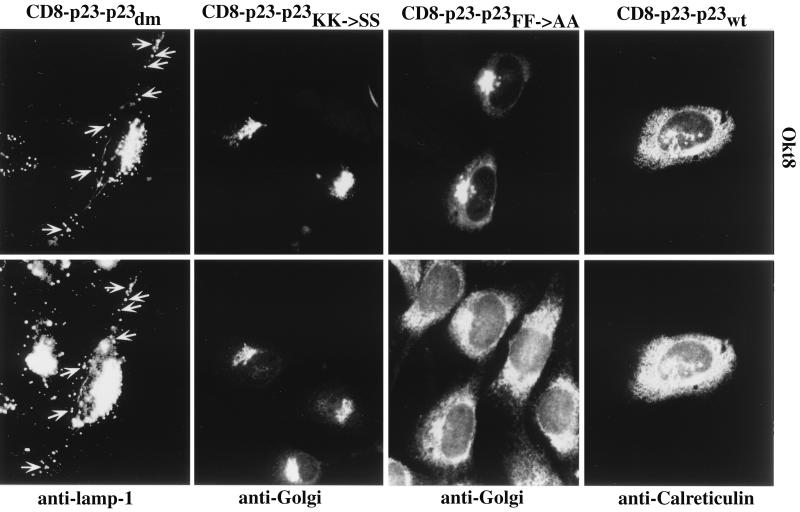
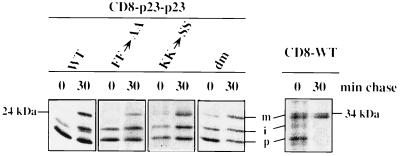
When the membrane-spanning domain and the cytoplasmic tail of p23 were transplanted to the lumenal domain of CD8 the resulting fusion protein (CD8–p23–p23wt) was directed to the ER (Fig. 2) as judged by colocalization with the ER marker protein calreticulin (25, 26). A minor fraction of transfected cells showed, in addition to ER staining, weak Golgi staining (data not shown). As endogenous p23 is predominantly localized to the Golgi complex (14) it can be concluded that this steady-state localization of p23 is neither determined by the membrane-spanning domain nor by its cytoplasmic tail but is dependent on its lumenal domain. However, the CD8–p23–p23wt fusion protein was capable to leave the ER as demonstrated by pulse–chase analysis (see Fig. 5). When transported to the IC/cis-Golgi area and the medial/trans-Golgi, the CD8 lumenal domain receives O-linked sugars giving rise to an intermediate and a mature form of the protein (24). After a 30-min pulse, both the nonglycosylated ER form as well as the intermediate form of CD8–p23–p23wt could be detected. After a 30-min pulse followed by a 30-min chase incubation, the mature form of CD8–p23–p23wt could also be detected (see Fig. 5). These data demonstrate that the fusion protein is transported out of the ER thereby receiving O-linked sugars. Because the construct is almost exclusively localized to the ER at steady-state, this localization appears to be maintained, at least in part, by active retrieval from the Golgi. The data are consistent with the interaction of the p23 tail with coatomer in vitro. Because the CD8–p23–p23wt fusion protein is efficiently retrieved to the ER it can be assumed that this process is based on an interaction of the p23 tail with COPI resulting in an efficient sorting of the molecule into retrograde transport vesicles. In contrast, a fusion protein composed of CD8 (lumenal domain) and the transmembrane domain as well as the cytoplasmic tail of p24 has been localized to the plasma membrane and the Golgi complex (16). Therefore, the cytoplasmic tail of p24 is not able to function as a retrieval signal when transplanted to a cell surface protein. Thus, p23 and p24 bear distinct intracellular sorting signals in their cytoplasmic tails.
Figure 2.
Transient expression of CD8–p23–p23 constructs in NRK cells. Cells were grown to about 30% confluence on coverslips. DNA transfer was by calcium phosphate precipitation. Twenty-four hours posttransfection cells were further processed for indirect immunofluorescence using the primary antibodies indicated. Arrows indicate some of the vesicular structures observed with the CD8–p23–p23dm construct that are colocalized with the lysosomal marker lamp-1. Note that here the perinuclear region has been overexposed to visualize peripheral lamp-1-positive structures.
Figure 5.
Pulse–chase analysis of CD8–p23–p23 fusion proteins. COS cells were grown on culture dishes up to 30% confluence and transfected with the various constructs using the calcium phosphate precipitation procedure. Twenty-four hours posttransfection the cells were pulse-labeled with [35S]methionine/cysteine and chased as described. At the times indicated the cells were kept on ice and lysed with a buffer containing Triton X-100. CD8–p23–p23 fusion proteins were immunoprecipitated from the various lysates using the monoclonal antibody OKT8 directed against the lumenal domain of CD8. Precipitates were separated on 12% SDS/polyacrylamide gels and analyzed by autoradiography. The unglycosylated precursor, the intermediate form, and the mature form of CD8 have been indicated as p, i, and m, respectively. Note that the absolute amounts of radioactivity incorporated into the various samples cannot be compared because individual samples correspond to individual culture dishes.
Mutations in the Coatomer Binding Motifs of the Cytoplasmic Tail of p23 Affect the Steady-State Localization of CD8–p23 Fusion Proteins.
In a previous study we had shown that coatomer binds to the cytoplasmic tail of p23 via two different motifs (14). First, a double phenylalanine motif in position −8/−9 was shown to be essential for coatomer binding. In addition, the tail contains a KKXX-related motif, KKLIE, at the extreme C terminus, which was shown to modulate coatomer binding. Coatomer binding was completely abolished when the FF motif was mutated even in the presence of the KK motif. On the other hand, coatomer binding was reduced only to about 50% of control levels when the KK motif was mutated. Therefore, the coatomer binding properties of the p23 tail are different from those of resident ER proteins carrying a classical KKXX retrieval signal where coatomer binding depends exclusively on the KKXX signal (7).
To analyze whether mutations in the coatomer binding motifs of the cytoplasmic tail of p23 affect the localization of CD8–p23 fusion proteins in vivo we constructed various versions with or without the membrane spanning domain of p23 (Fig. 1). As in the previous section, the results were compared with those obtained with a CD8–p24 fusion protein carrying an FF to AA mutation. Three mutants were constructed, either bearing an FF to AA mutation (CD8–p23–p23FF→AA), a KK to SS mutation (CD8–p23–p23KK→SS), or a double mutation in the cytoplasmic tail of the CD8–p23 fusion protein (CD8–p23–p23dm). Like CD8–p23–p23wt, all three mutant fusion proteins were shown to aquire O-linked sugars, indicating passage through the Golgi (see Fig. 5). A mutation in the FF motif resulted in a pronounced Golgi staining of the fusion protein although ER staining was still detectable (Fig. 2). Although the fusion protein seems to be predominantly localized to the Golgi, it might well be equally distributed between the Golgi and the ER because the signal is expected to become significantly weaker in a large compartment like the ER. These data indicate that retrieval from the Golgi is partially impaired by the FF to AA mutation. Likewise, replacement of the FF motif by AA in a CD8–p24 fusion protein results in a partial block of transport through the early secretory pathway or in a block of intra-Golgi transport (16).
The conversion of the KK motif to SS resulted in a complete abolishment of ER localization: the CD8–p23–p23KK→SS fusion protein was detected in the Golgi complex without any staining of the ER (Fig. 2). Thus, the KK motif is essential to confer ER localization to CD8–p23 fusion proteins. In summary, only with both coatomer binding motifs present the CD8–p23 fusion protein is strikingly retrieved to the ER. Disruption of both coatomer binding motifs (CD8–p23–p23dm) resulted in the appearance of the fusion protein in large vesicular structures mainly colocalizing with the lysosomal marker protein lamp-1 (27, 28) (Fig. 2). The peripheral vesicles positive for CD8–p23–p23dm were negative for the IC marker p58, the rat homologue of ERGIC53 (data not shown). The appearance of the fusion protein in vesicular structures of the endocytic pathway is possibly due to a LI motif at position −1/−2 that is known to mediate internalization of type I and type II membrane proteins into endosomal compartments when located in the cytoplasmic tail 10 or 20 amino acids distant from the lipid bilayer (29, 30). When the cells were not permeabilized before antibody incubation, a minor amount of CD8–p23–p23dm could also be detected at the cell surface (data not shown). These results indicate that retrograde transport of CD8–p23 fusion proteins from the Golgi depends on the presence of at least one coatomer binding motif. In a CD8–p24 construct, an FF to AA mutation impairs anterograde transport through the secretory pathway (16).
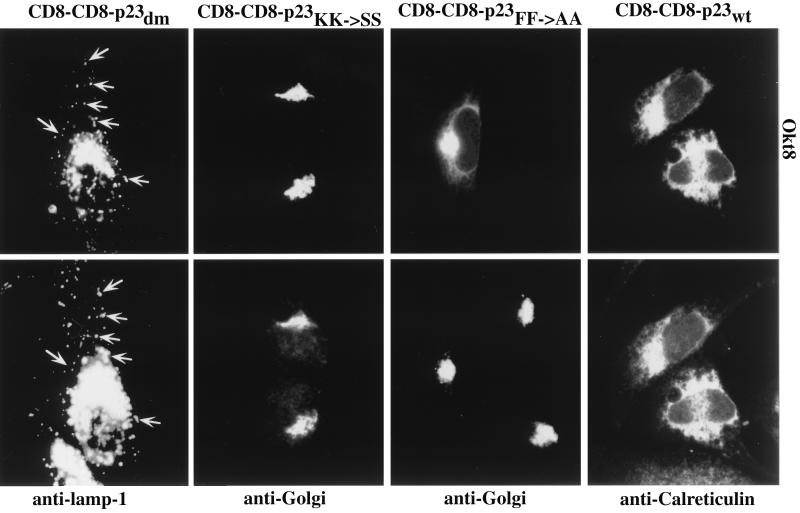
In all cases investigated the replacement of the p23 membrane span by the one of CD8 (CD8–CD8–p23 constructs: wild-type/FF→AA/KK→SS/double mutant) did not affect the steady-state localization of the fusion proteins (Fig. 3). Thus, the membrane-spanning domain does not appear to have a major impact on the subcellular localization of p23. Expression of the various CD8–p23 fusion proteins in HeLa and COS cells gave similar results compared with NRK cells (data not shown).
Figure 3.
Transient expression of CD8–CD8–p23 constructs in NRK cells. Cells were grown to about 30% confluence on coverslips. DNA transfer was by calcium phosphate precipitation. Twenty-four hours posttransfection cells were further processed for indirect immunofluorescence using the primary antibodies indicated. Arrows indicate some of the vesicular structures observed with CD8–CD8–p23dm construct that are colocalized with the lysosomal marker lamp-1. Note that here the perinuclear region has been overexposed to visualize peripheral lamp-1-positive structures.
Endogenous p23 Cycles Constitutively Through the Early Secretory Pathway.
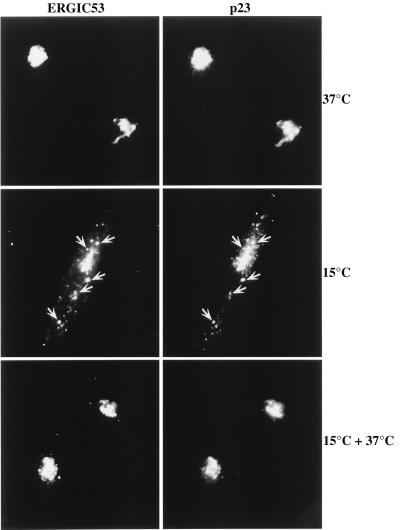
As outlined in the previous sections the cytoplasmic tails of p23 and p24 contain different intracellular sorting signals. These might be interpreted in terms of different intracellular routes these proteins may take. To further study the intracellular dynamics of p23 we performed temperature shift experiments using HeLa cells. To this end we used an affinity-purified polyclonal antibody directed against the cytoplasmic tail of p23. The appearance of p23 in the IC after incubating cells at 15°C was monitored by colocalization with the IC marker protein ERGIC53 (31, 32). The distribution of p23 was analyzed at 37°C (steady-state), after a 2-h incubation at 15°C and after release (30 min at 37°C) from the 2-h incubation at 15°C. Under each condition p23 was largely colocalized with ERGIC53 (Fig. 4). At 37°C a perinuclear staining was observed that corresponds to the Golgi and, possibly, to perinuclear elements of the IC. Peripheral elements of the IC were not stained. In contrast, at 15°C p23 accumulated in peripheral elements of the IC colocalized with ERGIC53. Release from the transport block resulted in a redistribution of p23 and ERGIC53 to the perinuclear area and a loss of staining of the peripheral IC elements. At present, there is no direct proof whether p23 recycles all the way back to the ER or whether it cycles between IC and Golgi. Although we were able to demonstrate that the various CD8–p23–p23 fusion proteins are able to leave the ER (Fig. 5), the rate of their transport is much slower when compared with the normal CD8 molecule (Fig. 5). Consistently, the ER-localized CD8–p23 fusion proteins (CD8–p23–p23wt and CD8–CD8–p23wt) did not dramatically redistribute when cells were incubated at 15°C (data not shown), similar to a CD4–ERGIC53 fusion protein (32) and a CD8–E19 fusion protein (23), and indicate that the signals in the cytoplasmic tails of recycling proteins are not sufficient for their continuous movement through the early secretory pathway. In contrast, it is the whole population of the endogenous p23 protein that cycles continuously between Golgi and IC and the cycling appears to depend both on the lumenal domain (because export of CD8–p23–p23wt/CD8–CD8–p23wt from the ER is relatively slow) and on its cytoplasmic tail that mediates retrieval from the Golgi. Based on immunofluorescence experiments, the cycling of p23 within the early secretory pathway is indistinguishable from the cycling of ERGIC53.
Figure 4.
Redistribution of endogenous p23 upon incubation of HeLa cells at 15°C. Cells were grown to about 80% confluence on coverslips. Before shifting the temperature the normal growth medium was replaced by Hepes-buffered medium to avoid pH changes during the incubation. The cells were either left at 37°C as a control, incubated for 2 h at 15°C, or incubated for 2 h at 15°C followed by a 30-min incubation at 37°C. After applying these conditions cells were further processed for indirect immunofluorescence using the primary antibodies indicated. Arrows indicate some of the peripheral IC elements after a 15°C incubation containing both ERGIC53 and p23.
DISCUSSION
This study is focused on the characterization of the intracellular dynamics of the COPI vesicle protein p23. By the replacement of the lumenal domain of p23 for the one of the cell surface protein CD8 it became possible to study sorting signals present in the membrane-spanning domain as well as in the cytoplasmic tail. We demonstrate that the cytoplasmic tail of p23 contains a functional retrieval signal that efficiently directs a CD8–p23 fusion protein to the ER. These data are consistent with an interaction of the p23 tail with COPI in vivo assuming that this retrieval is mediated by COPI-coated vesicles. Exchange of the transmembrane domain of p23 in all constructs did not affect the steady-state distribution of the corresponding fusion proteins and, therefore, these results suggest that it is the lumenal domain of p23 that determines its steady-state localization in the Golgi apparatus.
Mutations in the known coatomer binding motifs affect the steady-state localization of these fusion proteins within the ER–Golgi recycling pathway. Because all the CD8–p23–p23 fusion proteins are shown to be transported out of the ER to a significant extent it appears likely that the loss of coatomer binding motifs in the cytoplasmic tail results in a reduced ability to retrieve these fusion proteins from the Golgi back to the ER. Even more striking, constructs with tails that contain neither the FF nor the KK coatomer binding motif (CD8–p23–p23dm/CD8–CD8–p23dm) can escape from this pathway. Consistent with these observations, endogenous p23 is highly dynamic and cycles through the early secretory pathway. In contrast, another member of the p24 family, p24, was earlier shown not to contain a retrieval signal in its cytoplasmic tail (16). These data may be interpreted in terms of different routing of endogenous p23 and p24. On the other hand, it has been shown that their putative yeast homologues, Erv25p and Emp24p, form a heterodimer that is supposed to be important for COPII-mediated ER to Golgi transport. Furthermore, the turnover of Erv25p was shown to depend on the presence of Emp24p (15). Thus, it appears possible that mammalian p24 also cycles through the early secretory pathway depending on an interaction with p23. Mammalian p23 and p24 are clearly components of COPI-coated vesicles. Because their putative yeast homologues were localized to COPII-coated vesicles, it appears that the members of the p24 family are not specific to a certain class of COP-coated vesicles but are present in both COPI- and COPII-coated vesicles. If individual members of the p24 family exert specific roles in either anterograde or retrograde COPI vesicle transport, it remains to be established which is functional in a given vesicle or present solely as cargo.
A functional role of p23 as a Golgi-localized coatomer receptor has been proposed (14) based on the following criteria: p23 is a major membrane component of Golgi-derived COPI-coated vesicles present in amounts stoichiometric to coatomer and ADP ribosylation factor (ARF). Furthermore, p23 becomes concentrated into COPI-coated buds and vesicles upon addition to Golgi membranes, of cytosol and guanosine 5′-[γ-thio]triphosphate GTP[γS], and the cytoplasmic tail of p23 was demonstrated to bind coatomer in vitro. A second interaction of coatomer with the Golgi membrane is mediated by ARF which directly binds to β-COP in a GTP-dependent manner (33). This interaction is not restricted to the budding process but persists in purified COPI-coated vesicles. It appears, therefore, that coatomer binding to Golgi membranes is mediated by a bivalent interaction to p23 (and possibly other p24 proteins) and Golgi-bound ARF.
The average diameter of a COPI-coated vesicle is approximately 50 nm (inner diameter) and 70 nm (outer diameter), respectively. Thus, the coat is roughly 10 nm thick. The membrane surface of a 50-nm vesicle is about 7,600 nm2. If one coatomer complex covers an area of 10 × 10 nm then roughly 80 coatomer molecules form the coat of one vesicle. The stoichiometry between coatomer and p23 in a COPI-coated vesicle was found to be between 1:4 and 1:5 (14). Thus, one COPI-coated vesicle contains about 300–400 molecules of p23 (roughly 50,000 molecules/μm2). The spike proteins E1 and E2 of Semliki Forest virus, which replace the entire population of transported membrane proteins in infected cells and thus indicate an upper limit of cargo molecules, have a density of 1,700 molecules/μm2 of Golgi membrane (34). Therefore, not more than 3% of the membrane proteins of a COPI-coated vesicle are likely to represent cargo. From these calculations it appears that p24 proteins rather than KKXX-containing cargo proteins are the structural basis for coat binding during the formation of a COPI-coated vesicle. We assume that members of the p24 protein family form a scaffold that facilitates the polymerization of various COP coats. It is of note that some p24 family members like p23 contain a KKXX-related motif whereas others (i.e., p24) do not contain a double lysine motif. The cycling of p23 within the secretory pathway would be compatible with a functional role in both anterograde and retrograde transport. However, it is tempting to speculate that p23, with its conserved tail structure, forms the scaffold for retrograde moving COPI-coated vesicles. ER membrane proteins may have adopted the dilysine motif of p23 as a retrieval signal, enabling the few escaped copies present in the Golgi at any given time to become efficiently incorporated into the p23 scaffold, ensuring their sorting back to the ER. The intracellular dynamics of p24 will have to be established to obtain further insight into the function of these members of the p24 protein family.
Acknowledgments
We thank the members of the Wieland laboratory for discussions and comments on the manuscript. We are grateful to Dr. Hans-Dieter Söling (Göttingen, Germany), Dr. Walter Hunziker (University of Lausanne, Lausanne, Switzerland), and Dr. Hans-Peter Hauri (Biocenter Basel, Basel, Switzerland) for their kind gifts of antibodies directed against calreticulin, lamp-1, and ERGIC53, respectively. The eukaryotic expression plasmid pCMUIV containing the human CD8 cDNA was kindly provided by Dr. Tommy Nilsson (European Molecular Biology Laboratory, Heidelberg). This work was supported by grants from the Deutsche Forschungsgemeinschaft (to F.T.W.), the Human Frontiers Science Program Organization (to F.T.W.), and a fellowship from the Deutsche Forschungsgemeinschaft (to W.N.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ER, endoplasmic reticulum; IC, intermediate compartment; NSF, N-ethylmaleimide-sensitive fusion protein.
References
- 1.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 2.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 3.Pepperkok R, Scheel J, Horstmann H, Hauri H P, Griffiths G, Kreis T E. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- 4.Peter F, Plutner H, Zhu H, Kreis T E, Balch W E. J Cell Biol. 1993;122:1155–1167. doi: 10.1083/jcb.122.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bednarek S Y, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 6.Orci L, Glick B S, Rothman J E. Cell. 1986;46:171–184. doi: 10.1016/0092-8674(86)90734-8. [DOI] [PubMed] [Google Scholar]
- 7.Cosson P, Letourneur F. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 8.Letourneur F, Gaynor E C, Hennecke S, Demolliere C, Duden R, Emr S D, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 9.Schröder S, Schimmöller F, Singer K B, Riezman H. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis M J, Pelham H R B. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- 11.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach M F, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–908. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 12.Stamnes M A, Craighead M W, Hoe M H, Lampen N, Geromanos S, Tempst P, Rothman J E. Proc Natl Acad Sci USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schimmoeller F, Singer K B, Schroeder S, Krueger U, Barlowe C, Riezman H. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms J B, Wieland F T. J Cell Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belden W J, Barlowe C. J Biol Chem. 1996;271:26939–26949. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- 16.Fiedler K, Veit M, Stamnes M A, Rothman J E. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- 17.Harter C, Pavel J, Coccia F, Draken E, Wegehingel S, Tschochner H, Wieland F. Proc Natl Acad Sci USA. 1996;93:1902–1906. doi: 10.1073/pnas.93.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elrod-Erickson M J, Kaiser C A. Mol Biol Cell. 1996;7:1043–1058. doi: 10.1091/mbc.7.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trinchera M, Ghidoni R. J Biol Chem. 1989;264:15766–15769. [PubMed] [Google Scholar]
- 20.Jeckel D, Karrenbauer A, Birk R, Schmidt R R, Wieland F T. FEBS Lett. 1990;261:155–157. doi: 10.1016/0014-5793(90)80659-7. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson T, Jackson M, Peterson P A. Cell. 1989;58:707–18. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 22.Saraste J, Kuismanen E. Cell. 1984;38:535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- 23.Jackson M R, Nilsson T, Peterson P A. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascale M C, Erra M C, Malagolini N, Serafini C F, Leone A, Bonatti S. J Biol Chem. 1992;267:25196–25201. [PubMed] [Google Scholar]
- 25.Peter F, Nguyen V P, Söling H D. J Biol Chem. 1992;267:10631–10637. [PubMed] [Google Scholar]
- 26.Sönnichsen B, Füllekrug J, Nguyen V P, Diekmann W, Robinson D G, Mieskes G. J Cell Sci. 1994;107:2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
- 27.Lewis V, Green S A, Marsh M, Vihko P, Helenius A, Mellman I. J Cell Biol. 1985;100:1839–1847. doi: 10.1083/jcb.100.6.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green S A, Zimmer K P, Griffiths G, Mellman I. J Cell Biol. 1987;105:1227–1240. doi: 10.1083/jcb.105.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakke O, Dobberstein B. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 30.Pieters J, Bakke O, Dobberstein B. J Cell Sci. 1993;106:831–846. doi: 10.1242/jcs.106.3.831. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer A, Fransen J A, Bachi T, Ginsel L, Hauri H P. J Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itin C, Schindler R, Hauri H P. J Cell Biol. 1995;131:57–67. doi: 10.1083/jcb.131.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Helms J B, Brügger B, Harter C, Martoglio B, Graf R, Brunner J, Wieland F T. Proc Natl Acad Sci USA. 1997;94:4418–4423. doi: 10.1073/pnas.94.9.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinn P, Griffiths G, Warren G. J Cell Biol. 1994;98:2142–2147. doi: 10.1083/jcb.98.6.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]