Abstract
Podospora anserina is a filamentous fungus with a limited life span. Life span is controlled by nuclear and extranuclear genetic traits. Herein we report the nature of four alterations in the nuclear gene grisea that lead to an altered morphology, a defect in the formation of female gametangia, and an increased life span. Three sequence changes are located in the 5′ upstream region of the grisea ORF. One mutation is a G → A transition at the 5′ splice site of the single intron of the gene, leading to a RNA splicing defect. This loss-of-function affects the amplification of the first intron of the mitochondrial cytochrome c oxidase subunit I gene (COI) and the specific mitochondrial DNA rearrangements that occur during senescence of wild-type strains. Our results indicate that the nuclear gene grisea is part of a molecular machinery involved in the control of mitochondrial DNA reorganizations. These DNA instabilities accelerate but are not a prerequisite for the aging of P. anserina cultures.
Aging of biological systems, as the progressive functional impairment of living beings leading to an increase in age-related mortality, is a complex process that is dependent on both environmental and genetic factors. With regard to the genetic basis, instabilities of the mitochondrial DNA (mtDNA) were reported in various organisms to be involved in aging leading to mitochondrial dysfunction and degeneration (for reviews, see refs. 1 and 2). Moreover, increased instabilities of the mtDNA were found to lead to premature degeneration processes. Examples of this type are premature death syndromes in fungi and various neuromuscular diseases in humans (for reviews, see refs. 3–5). Interestingly, in some cases, these syndromes are due to the mutation of a nuclear gene that demonstrate that the integrity of the mtDNA is under the control of nuclear factors (6–10). Unfortunately, until now, the molecular mechanisms involved in this maintenance function have not been elucidated in any case.
In the filamentous fungus Podospora anserina, all wild-strains are characterized by a limited life span. After prolonged vegetative growth, they display various symptoms of senescence. Since the first description of this syndrome (11), it has become clear that nuclear–mitochondrial interactions are crucially involved in its control (for reviews, see refs. 12–15). Detailed molecular investigations revealed that gross reorganizations of the mtDNA occur during senescence. These reorganizations are almost quantitative and lead to impairment of the energy-generating system located in mitochondria. Similar processes were subsequently reported to occur in senescing strains of Neurospora (16–18). In all cases, the activity of mobile genetic elements of circular or linear structure were found to be relevant. More specifically, in P. anserina, it appears that the mobility of a circular plasmid, termed plasmid-like DNA (plDNA) or αsenDNA (19, 20), which is a derivative of the first intron of the cytochrome oxidase gene subunit I (COI), is responsible for the prominent age-related mtDNA rearrangements (21–23).
Because of experimental difficulties in the cloning of relevant nuclear genes, very little is known about the role these nuclear traits play in the control of senescence. In particular, the molecular interactions between age-related nuclear genes and mitochondrial genetic traits are unclear. Herein we report data of investigations into this direction. We focused our attention on P. anserina mutants displaying a long-lived phenotype because products of the wild-type copies of the corresponding genes appear to be involved in limiting life span as it occurs normally during aging. In contrast, short-lived mutants may rather be related to diverse pathologies of a given species (for further explanation of this concept, see ref. 24). Recently, we cloned the wild-type copy of the nuclear gene grisea that is mutated in this long-lived strain. The initial molecular characterization suggested that grisea codes for a copper-dependent transcription activator (15). This finding is in good agreement with the observed pleiotropic effects displayed by the long-lived mutant that was selected after chemical mutagenesis (25). We show that this mutant is a loss-of-function mutant in which the expression of grisea is defective. In addition, the amplification of plDNA and the wild-type-specific reorganizations of the mtDNA are affected. With respect to senescence, the data strongly suggest that, despite the rather unusual age-dependent reorganizations of the mtDNA occurring in wild-type strains of P. anserina, the basic mechanism(s) leading to senescence in this fungal model and in other biological systems appear(s) conserved but modulated by various factors.
MATERIALS AND METHODS
Strains and Media.
P. anserina wild-type strain s and the long-lived mutant grisea were used. For details of strain origin see refs. 25 and 26. Standard growth conditions for P. anserina were reported by Esser (26). For the discrimination of the wild-type and the grisea phenotypes, cornmeal agar was used. Special media used in transformation experiments and the detailed procedure to prepare and transform protoplasts of P. anserina were as described (27).
Plasmids.
Plasmids pKP230, pKP235, pKP240, pKP245 (28), pPSB2a, pPSB2b, pPSB4 (29), and pSP17 (plDNA) (30) were used as the source to isolate cloned mtDNA fragments of the wild-type strain s. These fragments were used as mtDNA-specific probes to detect age-dependent mtDNA reorganizations.
Plasmid pNP2-1 was used as an Escherichia coli/fungal shuttle vector to subclone parts of the grisea gene for complementation assays (15). In pPgr2-6, a 2.6-kb SmaI–NotI fragment containing the 5′ part of the wild-type grisea reading frame was subcloned (15). pPgr4-5 contains a 4.5-kb SmaI fragment derived from cosmid pPgr37-6 (15). The corresponding construct contains the entire reading frame of the wild-type allele of grisea. pPgr−6-7 contains a 6.7-kb NotI–ClaI fragment of the mutant allele of grisea. This fragment was isolated from a clone selected from a λ-EMBL3 genomic library of the mutant by hybridization with a wild-type probe of grisea. pPgrI−2-6 and pPgrP−6-7, respectively, contain hybrid sequences of the two grisea alleles. A 1.7-kb XbaI–NotI fragment of plasmid pPgr2-6 was exchanged with the corresponding sequence of the mutant gene copy derived from pPgr−6-7. The resulting construct, pPgrI−2-6, contains the wild-type sequence of grisea with the single G → A transition in the intron. Vice versa, the exchange of the mutant 1.7-kb XbaI–NotI fragment of pPgrP−6-7 with the corresponding wild-type fragment resulted in a hybrid plasmid, pPgr−6-7, that contains the three mutations in the 5′ upstream region of grisea but the wild-type intron sequence.
Construction of a Genomic λ Gene Bank and Selection of a grisea-Specific λ Recombinant Clone.
Total genomic DNA of long-lived mutant grisea was partially digested with Sau3AI and fractionated on agarose gels. DNA fragments between 16 and 23 kb were recovered and ligated into BamHI-digested EMBL3 arms (Stratagene). DNA packaging was performed by using Gigapack gold packaging extracts (Stratagene). All procedures for handling the λ gene bank and selection of recombinant clones carrying a grisea-specific insert were according to protocols provided by Stratagene or according to standard protocols (31).
cDNA Isolation and PCR Amplification.
For the isolation of cDNA, total RNA was first preamplified by using a (dT)12–16 primer and the SuperScript preamplification system from GIBCO/BRL. First-strand synthesis was performed according to the supplier recommendations. Subsequently, the cDNA was amplified by PCR using different oligonucleotide primer combinations. For the demonstration of the splicing defect in mutant grisea, un51 (5′-AGATGCCCATCATAAACG-3′) and un64 (5′-AACTGGTAGCACTGTCGA-3′) were used. These two 18-mers correspond to the positions 1367–1384 and 1669–1652 of the wild-type sequence of grisea (GenBank accession no. X89429).
PCR amplification was performed under the following conditions: denaturation for 2 min at 95°C and 30 amplification cycles (1 min, 95°C; 1 min, 57°C; 1 min, 72°C). The amplification products were fractionated on agarose gels and DNA fragments were recovered by using the Geneclean extraction kit (Bio 101).
Southern Blot Analysis.
In general, standard methods of molecular biology (e.g., bacterial transformation, DNA isolation, and Southern and Northern blot analyses) were performed according to Sambrook et al. (31). Southern blot analysis of DNA from Podospora cultures with different ages was performed by using the DIG detection system (Boehringer). Total DNA was isolated as described (32). Digested DNA was gel-fractionated and transferred to nylon membranes (HybondN, Amersham Buchler) by using a VacuGene XL vacuum blotting device (Pharmacia). After UV-cross-linking of the DNA, hybridization of specific probes labeled with digoxigenin-11-dUTP was carried out (33) at 68°C for 16 h in fresh prehybridization solution containing denatured probe at 20 ng/ml. Filters were washed twice at room temperature in 2× SSC/0.5% SDS for 5 min and twice at 75°C in 0.1× SSC/0.1% SDS for 30 min. Signal detection was performed by using the DIG detection kit as described (34).
DNA Sequencing and Sequence Analysis.
Both strands of the genomic copy of grisea were sequenced by using T7 polymerase (Pharmacia). In part, sequencing was performed after the generation of overlapping, sequentially shortened DNA fragments using a nested deletion kit (Pharmacia). The entire sequence of the mutant grisea allele was determined by using either the universal and reverse oligonucleotide primers or specific synthetic oligonucleotides. Sequence alterations in the mutant grisea allele will appear in the GenBank database under the accession number X89429.
RESULTS
Sequence Alterations in the Mutant Allele of grisea.
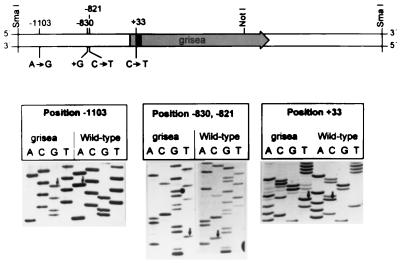
The wild-type copy of grisea was previously cloned by complementation of the corresponding long-lived mutant using a chromosome I-specific cosmid library of the wild-type strain (15). After an initial molecular characterization of this allele, we used a gene-specific probe to clone the mutant copy from a genomic λ library of the long-lived mutant. Subsequently, the nucleotide sequence of both strands was determined. Comparison of the corresponding sequence with the sequence of the wild-type allele (GenBank accession no. X89429) revealed three point mutations, two transitions, and a 1-bp addition, in the cloned genomic DNA at positions −1103, −830, and −821. In addition, a single G → A transition at the 5′ splice site of the single grisea intron was identified (Fig. 1).
Figure 1.
Localization of grisea on a 4.5-kb genomic SmaI fragment of P. anserina. The ORF of grisea is indicated by an arrow, the single intron is shown by the solid bar in this reading frame. The position and the nature of four sequence alterations in the mutant gene copy of grisea are indicated and correspond to the changes shown in the sequencing gels in the lower part of the figure. Arrows indicate three transitions and the dot one nucleotide addition. The numbers correspond to the positions of the corresponding nucleotides with respect to the the start codon of the grisea ORF (position +1).
RNA Splicing in Mutant grisea.
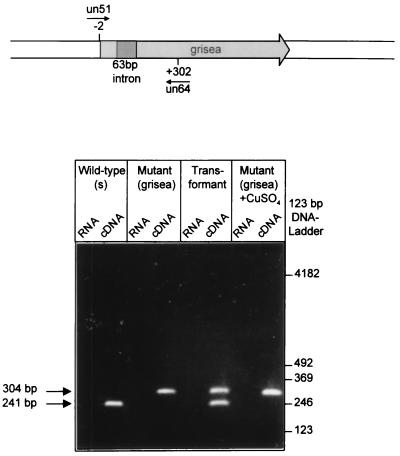
Because the first 5′ nucleotide of an intron is known to be involved in the RNA splicing reaction during gene expression, we surmised that the expression of grisea may be defective at this step. To verify this assumption, we analyzed total RNA from the wild-type of P. anserina, from the long-lived mutant, from a transformant in which the mutant phenotype was complemented by the introduction of the cloned wild-type copy of grisea, and from a mutant strain grown in the presence of additional copper. The latter analysis was performed because it was shown earlier that additional copper in the medium rescues the mutant phenotype (35). cDNA of the different strains was synthesized by using an oligo(dT) primer. Subsequently, the first-strand cDNA was amplified by PCR using specific oligonucleotide primer combinations that bind to sequences located upstream and downstream of the site of intron insertion in the grisea gene. In these experiments, cDNA from all strains led to specific amplification products. By using the oligonucleotides un51 and un64, a predominant product of approximately 240 bp was amplified (Fig. 2). This fragment exactly corresponds to the expected mature grisea-specific mRNA. In contrast, cDNAs derived from mutant grisea grown in medium with or without additional copper, led to a larger product of about 300 bp. We surmised that this amplification product is derived from the larger unspliced pre-mRNA of grisea. As expected, the cDNA derived from a transformant that displayed wild-type characteristics shows two major amplification products. One of these products corresponds to the spliced wild-type mRNA that is derived from the introduced wild-type transgene. The other band results from the amplification of the unspliced pre-mRNA and is a product of the resident mutant gene copy. In control experiments with DNase I-treated RNA of the different strains, no amplification products were obtained.
Figure 2.
Reverse transcription-coupled PCR analysis demonstrating a splicing defect in mutant grisea. RNA was isolated from the wild-type strain (s), from mutant grisea grown on medium with or without additional copper and from a transformant of grisea displaying wild-type characteristics due to an additional wild-type gene copy of grisea. Amplification was performed by using a specific pair of synthetic primers (un51 and un64) located upstream and downstream of the grisea intron. The four cDNA regions show the results from the PCR amplification after reverse transcription of RNA samples. Whereas the intron is almost completely spliced out of the pre-mRNA from the wild-type strain, no splicing product was detected in cDNA preparations of mutant grisea grown on medium with or without additional copper. The transformant gives rise to amplification products, 241 bp and 301 bp long, related to the spliced and unspliced RNA, respectively. The four RNA panels show the results of amplification of RNA prior to cDNA preparation (negative control).
To verify the nature of the amplification products, the 240-bp band of the wild-type strain and the 300-bp band of mutant grisea grown in standard medium were gel-eluted and sequenced. The nucleotide sequence verified the expected nature of the amplification products and the assumption that mutant grisea is a splicing deficiency mutant (data not shown). Furthermore, since a grisea-specific product from a reverse transcription-coupled PCR amplification can be identified also in assays using RNA from the mutant, the three point mutations located upstream of the deduced start codon of grisea, in a region that may be involved in the transcriptional control, do not lead to a complete block in the transcription of this gene.
Complementation Analyses.
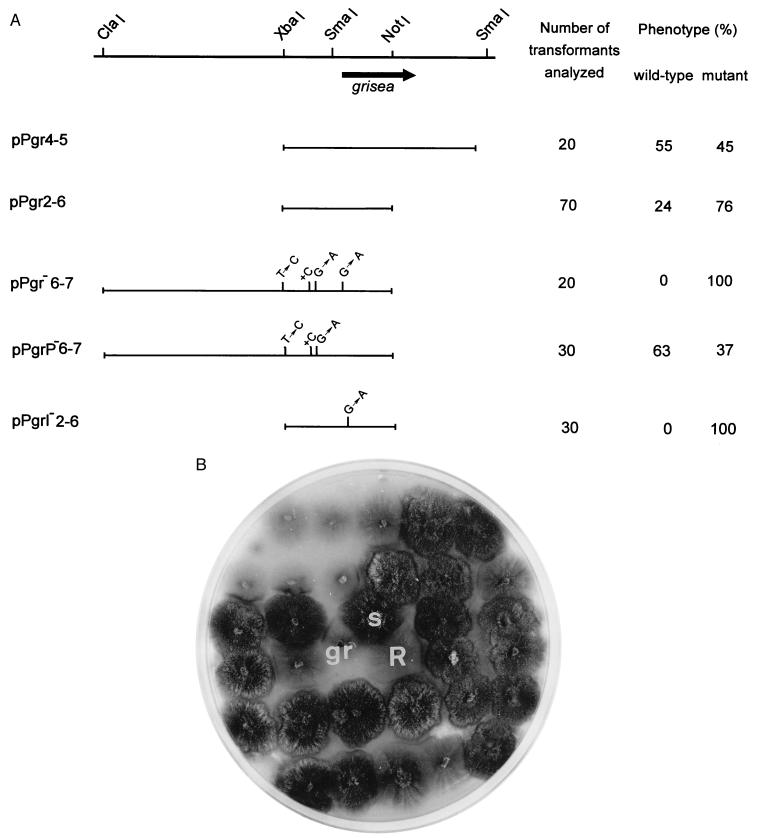
To further ascertain that it is the G → A transition identified in the mutant copy of grisea that is responsible for the mutant phenotype, we constructed various plasmids and used these constructs in transformation experiments to complement the mutant. As shown in Fig. 3, complementation of the mutant phenotype occurs not only with plasmids containing the whole grisea reading frame of the wild-type allele but also with constructs (pPgr2-6) in which only parts of the sequence coding for the amino terminus of the gene product are cloned. On the other hand, as expected, the complementation capacity was lost in a construct containing the same part of the grisea ORF as pPgr2-6 but that is derived from the mutant allele (pPgr−6-7). Most importantly, a chimeric construct (pPgrP−6-7) containing the three 5′ upstream mutations but the wild-type sequence at the 5′ splice site of the grisea intron was able to complement the mutant phenotype (Fig. 3B). In contrast, the introduction of the G → A transition into a cloned (pPgrI-2-6) DNA fragment that does not contain the three mutations upstream of the start codon resulted in a loss of complementation capacity. We conclude that it is the intron mutation that is responsible for the observed phenotype of long-lived mutant grisea.
Figure 3.
Complementation assay demonstrating that a mutation in the single grisea intron leads to a loss of function. (A) A physical map of the genomic grisea locus is presented at the top. Various plasmids were constructed and used to complement long-lived mutant grisea to wild-type characteristics. In plasmids pPgr−6-7, pPgrP−6-7, and pPgrI−2-6, point mutations corresponding to the mutations identified in the mutant grisea allele are indicated. The right column summarizes the results of the transformation experiments. (B) Morphology of the untransformed recipient strain grisea (gr), the wild-type strain s (s), a colony regenerated from protoplasts of grisea that were not incubated with DNA of plasmid pPgrP−6-7 (R), and independent hygromycin B-resistant transformants of grisea. Sixty-three percent of the transformants (dark colonies) show the typical wild-type morphology.
Age-Dependent mtDNA Reorganizations.
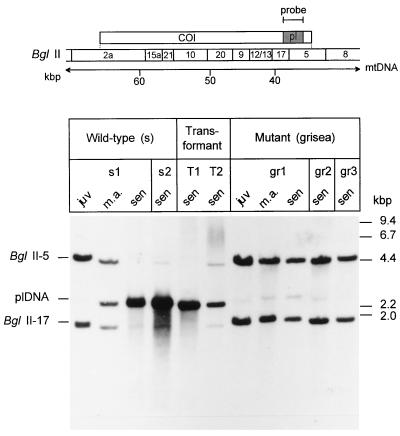
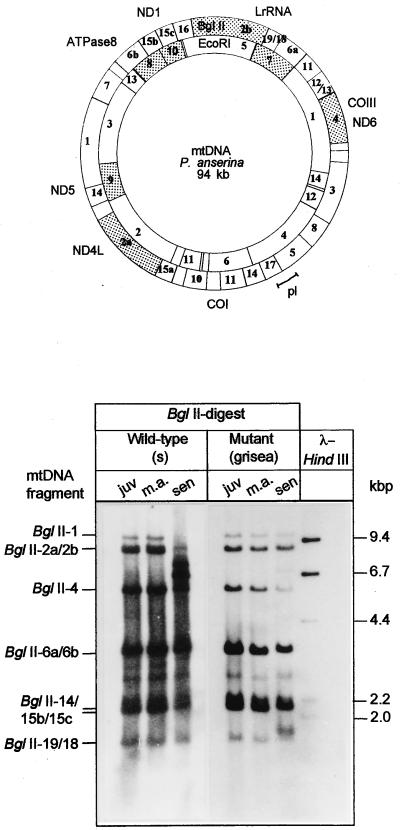
It is well documented that gross mtDNA rearrangements regularly appear during senescence of P. anserina cultures (36–38). To investigate a potential role of grisea in the control of these age-related mtDNA rearrangements, we analyzed DNA of the wild-type, the long-lived mutant grisea, and cultures of grisea that were transformed with the cloned wild-type copy of grisea and displayed wild-type characteristics. Integration of the transforming hybrid plasmid into the genomic DNA was verified by Southern blot analyses (data not shown). These experiments revealed that the vector containing the transgene was inserted into an ectopic site. The analysis of the mtDNA is shown in Fig. 4. Because of a single BglII restriction site in the pl-intron, two BglII fragments, BglII-5 and BglII-17, hybridize to the plDNA-specific probe (pSP17). These two fragments correspond to the pl-intron and adjacent mtDNA sequences. In juvenile cultures of the wild-type strain, the BglII-5 and BglII-17 fragments are the two major bands observed in Southern blots. Only a very faint 2.5-kb fragment corresponding to the linearized autonomous intron derivative (plDNA) can be detected. The relative amount of this fragment increases as cultures age. At the same time, the hybridization signals corresponding to the intact wild-type mtDNA strongly decrease. These data were in accordance with results from earlier investigations that demonstrate that gross mtDNA reorganizations occur during senescence of P. anserina wild-type strains. Unexpectedly, only trace amounts of the autonomous plDNA were identified in senescent cultures of the long-lived mutant. In addition, the intensity of the restriction fragments of the mtDNA that contain the pl-intron (fragments BglII-5 and BglII-17) did not change in DNA preparations from cultures of different age. Importantly, transformation of grisea protoplasts with the wild-type copy of grisea resulted in a reversion of this effect.
Figure 4.
Southern blot analysis of the plDNA region of mtDNA from different P. anserina cultures. Total DNA digested with BglII was fractionated on an agarose gel and hybidized with a plDNA-specific probe. The localization of the probe (pl) in the mtDNA region of the cytochrome oxidase subunit I gene (COI) and a BglII restriction map is shown in the scheme. DNA was isolated from independent cultures of the wild-type strain race s (s1, s2), long-lived mutant grisea (gr1, gr2, gr3), and grisea transformants (T1, T2) that, due to the transformation with the grisea wild-type gene copy, are complemented to wild-type characteristics. DNA was derived from juvenile (juv), middle aged (m.a.), and senescent (sen) cultures, respectively.
To extend the comparative analyses of age-related mtDNA rearrangements, we hybridized BglII-digested mtDNA from cultures of different age against different mtDNA-specific probes. These probes were derived from regions of the mtDNA that are not overlapping the region containing the pl-intron (28, 29). As shown in Fig. 5, during aging the two largest BglII fragments of the wild type become rearranged. Interestingly, these fragments remain stable in cultures of the mutant. However, relative to the other BglII fragments, the BglII-4 fragment of the mutant declines during aging and a new band appears in the region above the BglII-18/19 fragments. Thus, it appears that the nuclear encoded transcription factor GRISEA is somehow involved in the control of amplification of plDNA and in the wild-type-specific mtDNA instabilities that occur during senescence.
Figure 5.
Southern blot analysis of the P. anserina mtDNA regions that do not overlap the plDNA region. (Upper) Locations of the cloned DNA probes used in the hybridization experiments are shaded. (Lower) Hybridization experiments. Fragment numbers are according to ref. 28. Total DNA was derived from juvenile (juv), middle aged (m.a.), and senescent (sen) cultures of the wild-type and mutant grisea. DNA was digested with BglII and fractionated on an agarose gel.
DISCUSSION
Herein we report data of the molecular characterization of the long-lived mutant grisea of P. anserina. We have identified four point mutations in the cloned grisea locus and have demonstrated that one of them, a G → A transition at the 5′ splice site of the single grisea intron, is responsible for the mutant phenotype. This mutation leads to a RNA splicing defect. Thus, grisea is a loss-of-function mutant in which the expression of the corresponding gene is affected at the RNA splicing level.
In regard to senescence, the finding that grisea affects the amplification of the pl-intron and the age-related reorganization of the mtDNA is most striking. The data indicate that the liberation and/or the amplification of the plDNA is somehow under the control of this gene. Previously, it was suggested (23) that the liberation of plDNA proceeds via the transposition of the pl-intron leading to a tandem organization of intron copies in the mtDNA. Subsequent recombination between homologous sequences may lead to the generation of the circular plDNA. In addition to a reverse transcriptase that is encoded by the pl-intron (39), we suggest that one or a few gene product(s) that are expressed under the control of GRISEA may be involved in the process.
The results suggesting this scenario were rather unexpected because the amplification of plDNA was thought to be crucial for the induction of senescence (22, 36, 37) and should, therefore, also occur in mutant grisea. The analysis reported herein could not verify this assumption but indicates that the gross age-related mtDNA rearrangements reported in wild-type strains of P. anserina lead to an acceleration of the processes leading to senescence. In accordance with this view, an extrachromosomal mutant in which a delay in the amplification of the mobile intron was previously shown to be correlated with an increased life span (40). However, in this extrachromosomal mutant, this effect is not the result of the mutation in a nuclear gene but instead is due to the presence of a linear mtDNA plasmid (41, 42). Interestingly, recently the characterization of two nuclear mutants of P. anserina revealed that, like in mutant grisea, a defect in the amplification of plDNA does not prevent senescence. In the corresponding mutants genes encoding cytoplasmic ribosomal proteins are affected (43). Moreover, the characterization of another nuclear mutant of P. anserina displaying an early growth arrest (premature death) was reported to be characterized by the occurrence of a site-specific deletion of the mitochondrial genome (7, 8). In this mutant, two nuclear genes interact with each other. One of these genes, AS1-4, was found to code for a cytoplasmic ribosomal protein (44). The nature of the second gene, rmp, is still unclear. The same holds true for a nuclear gene, nd, that leads to increased mtDNA deletions between direct repeats and, as a result, to premature death in Neurospora crassa cultures (6).
Thus, it appears that a number of different genetic traits, either nuclear or mitochondrial, have an impact on both mtDNA reorganizations and on the life span of P. anserina. Moreover, the extensive age-related mtDNA rearrangements that are characteristic for the various wild-type strains of this fungus but that are not observed in other biological systems in such an dramatic form do not appear to be a prerequisite for aging. Another mechanism seems to operate also in P. anserina, a mechanism that may be conserved in the different biological systems. As suggested by others (45), such a mechanism may be related to different stresses (e.g., oxidative stress, heat shock, or cold stress) and the ability of the organism to cope with these situations. Until now, this is the only obvious relationship between a number of long-lived mutants that have been characterized.
The relevance of oxidative stress and the free radical theory of aging (46) for P. anserina is supported by the observation that grisea codes for a transcription factor that appears to be involved in a tight regulation of cellular copper levels and the fact that the phenotype of grisea can be rescued by additional copper in the growth medium (35). The increased life span of the mutant strain may be explained by a reduction in the formation of reactive oxygene species (ROS). Two reasons appear to account for this effect. (i) A decrease in ROS formation may be due to the absence of a high-affinity transporter for copper due to a reduced expression of the corresponding gene. Intracellular copper levels, therefore, may be low and ROS formation via Fenton chemistry is reduced (47). The addition of high amounts of copper to the medium may lead to an increased copper uptake that is independent from the suggested high-affinity transporter. Subsequently, increased levels of ROS occur and life span is shortened. This conclusion is in accordance with the data described for a mutant of Saccharomyces cerevisiae in which MAC1, a GRISEA homolog, is affected. The phenotype of this yeast mutant can also be rescued by additional copper in the medium. Moreover, it was found that the expression of FRE1, a gene that is involved in both iron and copper uptake (48, 49) is impaired. (ii) In the mutant grisea, another source of ROS formation appears to be greatly affected. This source is located in the mitochondrion and is dependent on the reorganization of the mtDNA. For humans it was suggested that increased numbers of defective mtDNA molecules lead to an impairment of the mitochondrial electron transport chain leading to the generation of increased levels of ROS and consequently to a severe molecular damage of various biomolecules (50). Because in the mutant grisea, as shown in this paper, the reorganization of the mtDNA appears to be reduced this source of ROS generation may be decreased.
Thus, the suggested scenario provides further clues about the details of a rather complex molecular network leading to senescence in P. anserina. In this scenario, genetic factors encoded by the nucleus and by mitochondria are involved in the genetic control of aging. Further progress in this field of experimental aging research requires the identification and characterization of the different target genes of the suggested transcription apparatus. Currently, different approaches into this direction are underway in our laboratory. The results of these investigations should certainly contribute to understanding the relevant molecular mechanisms in this fungal model system and should shed more light on mechanisms involved in the normal and the accelerated degeneration processes (e.g., aging, degenerative diseases) occurring in the various biological systems.
Acknowledgments
We are grateful to Drs. J. Hermanns and T. Schwartz (Heidelberg) for experimental help and useful suggestions. We thank Dr. T. Elthon (University of Nebraska, Lincoln, NE) for critical reading of the manuscript. The skillful technical assistance of Ms. A. Werner (Frankfurt, Germany) is gratefully acknowledged. The experimental work was performed within the framework of the Concerted Action Program (BMH1-CT94-1710) of the European Community and was supported by a grant of the Deutsche Forschungsgemeinschaft (Bonn-Bad Godesberg, Germany). Part of the work was performed at the German Cancer Research Center (Heidelberg).
ABBREVIATIONS
- plDNA
plasmid-like DNA
- ROS
reactive oxygen species
- mtDNA
mitochondrial DNA
Footnotes
Data desposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. X89429).
References
- 1.Griffiths A J F. Annu Rev Genet. 1992;26:351–372. doi: 10.1146/annurev.ge.26.120192.002031. [DOI] [PubMed] [Google Scholar]
- 2.Osiewacz, H. D. (1997) J. Mol. Med., in press.
- 3.Wallace D. Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 4.Osiewacz H D, Hermanns H D. Aging Clin Exp Res. 1992;4:273–286. doi: 10.1007/BF03324108. [DOI] [PubMed] [Google Scholar]
- 5.Osiewacz H D. In: Molecular Gerontology. Rattan S I S, Toussaint O, editors. New York: Plenum; 1996. pp. 37–52. [Google Scholar]
- 6.Seidel-Rogol B, King J, Bertrand H. Mol Cell Biol. 1989;9:4259–4264. doi: 10.1128/mcb.9.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belcour L, Begel O, Picard M. Proc Natl Acad Sci USA. 1991;88:3579–3583. doi: 10.1073/pnas.88.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contamine V, Lecellier G, Belcour L, Picard M. Genetics. 1996;144:541–555. doi: 10.1093/genetics/144.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. Nature (London) 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 10.Suomalainen A, Kaukonen J, Amati P, Timonen R, Haltia M, Weissenbach J, Zeviani M, Somer H, Peltonen L. Nat Genet. 1995;9:146–151. doi: 10.1038/ng0295-146. [DOI] [PubMed] [Google Scholar]
- 11.Rizet G. C R Acad Sci. 1953;237:838–855. [PubMed] [Google Scholar]
- 12.Esser K. In: The 1984 Sandoz Lectures in Gerontology. Bergener M, Ermini M, Stähelin H B, editors. London: Academic; 1984. pp. 3–20. [Google Scholar]
- 13.Kück U. Exp Mycol. 1989;13:111–120. [Google Scholar]
- 14.Osiewacz H D. In: Molecular Aspects of Aging. Esser K, Martin G M, editors. New York: Wiley; 1994. pp. 29–44. [Google Scholar]
- 15.Osiewacz H D, Nuber U. Mol Gen Genet. 1996;252:115–124. doi: 10.1007/BF02173211. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand H, Griffiths A J F, Court D A, Cheng C K. Cell. 1985;41:877–884. doi: 10.1016/s0092-8674(85)80068-4. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand H, Griffiths A J F, Court D A, Cheng C K. Cell. 1986;47:829–837. doi: 10.1016/0092-8674(86)90525-8. [DOI] [PubMed] [Google Scholar]
- 18.Akins R A, Kelly R L, Lambowitz A M. Cell. 1986;47:505–516. doi: 10.1016/0092-8674(86)90615-x. [DOI] [PubMed] [Google Scholar]
- 19.Stahl U, Lemke P A, Tudzynski P, Kück U, Esser K. Mol Gen Genet. 1978;178:639–646. doi: 10.1007/BF00337872. [DOI] [PubMed] [Google Scholar]
- 20.Cummings D J, Belcour L, Grandchamps C. Mol Gen Genet. 1979;171:239–250. doi: 10.1007/BF00267578. [DOI] [PubMed] [Google Scholar]
- 21.Osiewacz H D, Esser K. Curr Genet. 1984;237:1–8. doi: 10.1007/BF00419728. [DOI] [PubMed] [Google Scholar]
- 22.Kück U, Osiewacz H D, Schmidt U, Kappelhoff B, Schulte E, Stahl U, Esser K. Curr Genet. 1985;9:373–382. doi: 10.1007/BF00421608. [DOI] [PubMed] [Google Scholar]
- 23.Sellem C H, Lecellier G, Belcour L. Nature (London) 1993;366:176–178. doi: 10.1038/366176a0. [DOI] [PubMed] [Google Scholar]
- 24.Fleming J E, Rose M R. In: Handbook of the Biology of Aging. Schneider E L, Rowe J W, editors. New York: Academic; 1996. pp. 74–93. [Google Scholar]
- 25.Prillinger H, Esser K. Mol Gen Genet. 1977;156:333–345. doi: 10.1007/BF00267190. [DOI] [PubMed] [Google Scholar]
- 26.Esser K. In: Handbook of Genetics. King R C, editor. New York: Plenum; 1974. pp. 531–551. [Google Scholar]
- 27.Osiewacz H D, Skaletz A, Esser K. Appl Microbiol Biotechnol. 1991;35:38–45. doi: 10.1007/BF00180633. [DOI] [PubMed] [Google Scholar]
- 28.Kück U, Esser K. Curr Genet. 1982;5:143–147. doi: 10.1007/BF00365705. [DOI] [PubMed] [Google Scholar]
- 29.Schulte E, Kück U, Esser K. Mol Gen Genet. 1988;211:342–349. [Google Scholar]
- 30.Stahl U, Tudzynski P, Kück U, Esser K. Proc Natl Acad Sci USA. 1982;79:3641–3645. doi: 10.1073/pnas.79.11.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Hermanns J, Asseburg A, Osiewacz H D. Curr Genet. 1995;27:379–386. doi: 10.1007/BF00352108. [DOI] [PubMed] [Google Scholar]
- 33.Höltke H I, Sagner G, Kessler C, Schmitz G. BioTechniques. 1992;12:104–113. [PubMed] [Google Scholar]
- 34.Düring K. Anal Biochem. 1991;19:433–438. doi: 10.1016/0003-2697(91)90490-k. [DOI] [PubMed] [Google Scholar]
- 35.Marbach K, Fernandez-Larrea J, Stahl U. Curr Genet. 1994;26:184–186. doi: 10.1007/BF00313809. [DOI] [PubMed] [Google Scholar]
- 36.Kück U, Stahl U, Esser K. Curr Genet. 1981;3:151–156. doi: 10.1007/BF00365719. [DOI] [PubMed] [Google Scholar]
- 37.Belcour L, Begel O, Mosse M-O, Vierny C. Curr Genet. 1981;3:13–21. doi: 10.1007/BF00419575. [DOI] [PubMed] [Google Scholar]
- 38.Wright R M, Horrum M A, Cummings D J. Cell. 1982;29:505–515. doi: 10.1016/0092-8674(82)90167-2. [DOI] [PubMed] [Google Scholar]
- 39.Faβbender S, Brühl K H, Ciriacy M, Kück U. EMBO J. 1994;13:2075–2083. doi: 10.1002/j.1460-2075.1994.tb06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osiewacz H D, Hermanns J, Marcou D, Triffi M, Esser K. Mutat Res. 1989;219:1–7. doi: 10.1016/0921-8734(89)90036-2. [DOI] [PubMed] [Google Scholar]
- 41.Hermanns J, Osiewacz H D. Curr Genet. 1992;22:491–500. doi: 10.1007/BF00326415. [DOI] [PubMed] [Google Scholar]
- 42.Hermanns J, Asseburg A, Osiewacz H D. Mol Gen Genet. 1994;243:297–307. doi: 10.1007/BF00301065. [DOI] [PubMed] [Google Scholar]
- 43.Silar P, Koll F, Rossignol M. Genetics. 1997;145:697–705. doi: 10.1093/genetics/145.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dequard-Chablat M, Sellem C H. J Biol Chem. 1994;21:14951–14956. [PubMed] [Google Scholar]
- 45.Martin G M, Austad S N, Johnson T E. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 46.Harman D. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 47.Halliwell B, Gutteridge M C. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jungmann J, Reins H-A, Lee J, Romeo A, Hassett R, Kosman D, Jentsch S. EMBO J. 1993;12:5051–5056. doi: 10.1002/j.1460-2075.1993.tb06198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dancis A, Yuan D S, Halle D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner R D. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- 50.Bandy B, Davison A J. Free Radical Biol Med. 1990;8:523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]