Abstract
Mutations in the nubbin (nub) gene have a phenotype consisting of a severe wing size reduction and pattern alterations, such as transformations of distal elements into proximal ones. nub expression is restricted to the wing pouch cells in wing discs since early larval development. These effects are also observed in genetic mosaics where cell proliferation is reduced in all wing blade regions autonomously, and transformation into proximal elements is observed in distal clones. Clones located in the proximal region of the wing blade cause in addition nonautonomous reduction of the whole wing. Cell lineage experiments in a nub mutant background show that clones respect neither the anterior–posterior nor the dorsal–ventral boundary but that the selector genes have been correctly expressed since early larval development. The phenotypes of nub el and nub dpp genetic combinations are synergistic and the overexpression of dpp in clones in nub wings does not result in overproliferation of the surrounding wild-type cells. We discuss the role of nub in the wing’s proximo–distal axis and in the formation of compartment boundaries.
Keywords: proximo–distal axis, accommodation, morphogenesis, compartment boundaries
In Drosophila melanogaster, the imaginal discs, which will give rise to adult structures, are subdivided into compartments (1). Since the beginning of embryogenesis, a segregation of anterior (A) and posterior (P) cells takes place and in the larval period a dorsal–ventral (D–V) restriction boundary is created in wing and haltere discs. As a consequence, cells from the A compartment do not intermix with cells of the P compartment from the blastoderm stage onward and cells in the D compartment do not cross over to the V compartment after the second larval stage. Moreover, the selector genes (those conferring compartment identity) (2) for these compartments are known. Expression of engrailed (en) is the definitive feature of P cells (3–7) and expression of apterous (ap) is characteristic of D cells (8). The A cells are the non-en-expressing cells, and the V cells are the cells that do not express ap. The regions of contact between D and V cells, and between A and P cells, the boundaries, are of special interest. The confrontation of cells from two different compartments leads to the expression of new genes in the boundaries, some of which are directly involved in cell proliferation (7, 9–13). In addition to A–P and D–V axes, there is another axis, the proximal–distal axis in adult appendages. The growth and patterning along this axis play an important role in the final shape and size of the organ. Some genes have been proposed to be crucial for the proximal–distal axis establishment in the legs, such as Distalless (Dll) (14, 15). Among the mutations affecting the growth and patterning of the proximal–distal axis of the wing we have chosen to study nubbin (nub), a gene encoding a putative transcription factor with a POU domain named pdm-1 (16). pdm-1 is expressed in a dynamic segmental pattern during embryonic development and is also expressed in the peripheral and central nervous systems (17–19). In this paper we demonstrate that the nub gene is important for correct proximal–distal specification of the growing wing imaginal disc. Moreover, we present evidence that incorrect specification of this axis due to mutations in nub leads to failures in growth along both the D–V and A–P axes.
MATERIALS AND METHODS
Fly Stocks.
All the mutants used in this study are as described (20). We have used two adult viable alleles, nub1 and nub2, a deficiency with a dominant nub phenotype Df(2L)Prl and a lethal deficiency Df(2L)prd.1.7.
Clonal Analysis.
Mitotic recombination was induced by x-rays (dose = 1,000 R; 300 R/min, 100 kV, 15 mA, 2-mm aluminum filter; 1 R = 0.258 m⋅C/kg). Clonal analysis was carried out for nub1, nub2, and Df(2L)Prl. Three different types of clonal analysis were done, M+ clones, twin analysis, and cell lineage. For M+ mosaics, a chromosome carrying a Minute mutation [M(2L)24F] and a f+ duplication located at 30B was used. nub/M(2L)24F f+30B larvae were irradiated at different ages, and adult wings were scored to find nub− cells marked with f. For twin analysis nub/f+30B ck pr pwn larvae were irradiated. For cell lineage analysis of the nub mutants, nub2; mwh/+ larvae were irradiated at 60 h after egg laying (AEL). We have also carried out a cell lineage study of M+ clones in nub2; mwh/M(3)67C larvae irradiated at different ages.
Imaginal Disc Staining.
In situ hybridizations, acridine orange, biotinilated antibody, and fluorescent antibody stainings were performed as described (21–24), respectively.
Tubα1>dpp Clones in nub Wings.
f36a; nub Tubα1>f+>dpp/nub; hsp70-flp (> = flipase recombinase target site) flies were generated and subjected to a heat shock (35°C for 30 min).
RESULTS
nub Phenotypes.
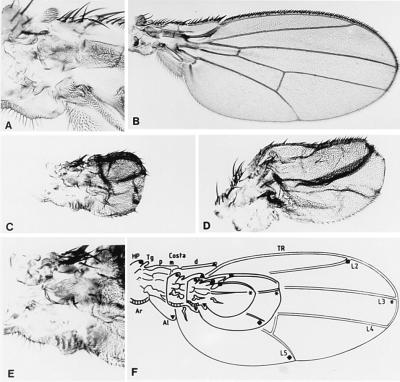
We have used two hypomorphic alleles, nub1 and nub2, nub1 being more extreme than nub2 (Fig. 1). Mutant wings are smaller and shorter than normal and are abnormally folded and bent. This wing size reduction is due to a reduction in the number of wing cells as demonstrated by normal trichome density. The bristles of the triple row are incorrectly differentiated, and dorsal and ventral elements are not easily distinguishable. In some cases bracteated bristles, normally located in the proximal costa, can be seen in distal positions, indicating a failure in proximo–distal specification. In nub2 the longitudinal vein L4 is absent and in nub1 both posterior longitudinal veins, L4 and L5, are missing. The crossveins are absent in both alleles. Notum and axillar patterns however are unaffected (Fig. 1 A and E). The haltere is smaller than wild type. The legs are not affected in nub2 allele but in nub1 may appear shortened and gnarled. These phenotypes are severely increased in a Minute background. The dominant haploinsufficient phenotype of Df(2L)Prl consists of a slight reduction in wing blade size and a lack of the vein L5.
Figure 1.
nub phenotypes. (A) Axillar region of a wild-type wing (B). (C) nub1 mutant wing. (D) nub2 mutant wing. (E) Axillar region of a nub1 mutant wing. (F) Comparison of wild-type and nub (the smaller of the two mutant wings is nub1 and the other is nub2) mutant wing patterns; L2-L5 are the longitudinal veins. HP, humeral plate; Tg, Tegula; Costa p, m, and d, Costa proximal, medial, and distal, respectively; TR, triple row; Ar, Arc; Al, Alula. The various symbols facilitate the comparison of wild-type and mutant patterns.
Cell Death.
To study cell mortality in nub mutants, wing discs were stained with acridine orange at different stages of larval development. In second larval instar discs, we have not detected cell death. In third larval instar mutant wings, we see conspicuous cell mortality mainly in the axillar region surrounding the wing pouch but not in the rest of the wing disc in a higher degree than in wild-type discs (Fig. 2A).
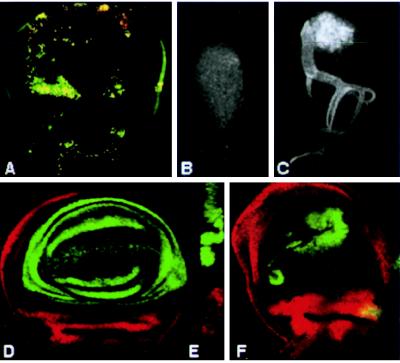
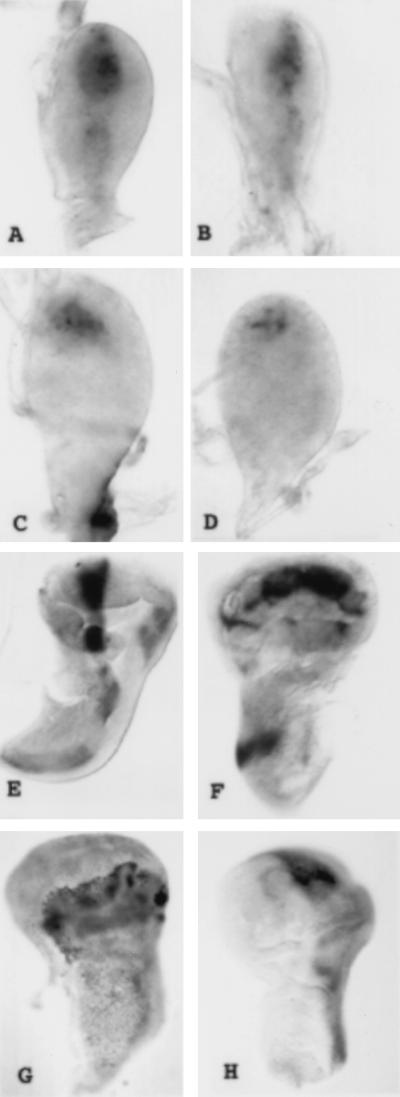
Figure 2.
Cell death and Pdm-1 expression. (A) Acridine orange staining showing the cell death pattern in a mature nub1 mutant wing disc. (B) Early second larval instar wild-type wing disc with no Pdm1 staining. (C) Anti-Pdm1 staining in 60-h-old second larval instar wild-type wing disc. (D–F) Confocal micrographs of wing imaginal discs doubly stained for Pdm-1 (green) and Teashirt (red). (D) Wild-type third larval instar wing imaginal disc showing the Pdm-1 staining restricted to the wing pouch. (E) Orthogonal section of the same wing disc that confirms the absence of coexpression of Pdm-1 and Teashirt. (F) Pdm-1 expression in nub1 mutant wing imaginal disc. No transformation to Teashirt expressing cells is observed in the wing pouch cells that do not express Pdm-1.
Pdm-1 Expression.
The pattern of protein expression was monitored with antibodies against Pdm-1. We are first able to detect the protein in around 60-h-old wing discs with between 150 and 200 cells. The protein is present in a subset of 30–40 cells in a region that probably gives rise to the adult wing blade (Fig. 2 B and C and ref. 25). In third larval stage wing imaginal discs, Pdm-1 protein is accumulated in the wing blade (Fig. 2D). These results are in accordance with pdm-1 RNA expression pattern (16). To characterize the wing-specific expression of pdm-1 in greater detail, Pdm-1–Teashirt double antibody stainings were carried out. The teashirt gene codes for a protein that is present in the notum and in the proximal region of the hinge (ref. 25 and data not shown). In double staining with antibodies against Pdm-1 and Teashirt, no cell shows coexpression of Teashirt and Pdm-1 and a group of cells that express neither protein separates the Pdm-1 and Teashirt domains (Fig. 2 D and E). This indicates that Pdm-1 is present only in the wing blade and not in the hinge, in accordance with nub mutant phenotypes. At the end of larval development the anti-Pdm-1 staining is stronger in the putative wing vein precursor cells (data not shown). During pupal development intervein staining decreases and in 24 h after puparium formation, old pupal wings Pdm-1 protein is observed only in stripes (6–8 cells wide) corresponding to the veins and in the basal trunks of the veins (J. Felix de Celis, personal communication). In nub mutant discs, we have detected Pdm-1 protein in a few wing pouch cells, indicating that the nub alleles used are not protein nulls. The wing pouch cells that do not express Pdm-1 are not expressing Teashirt, excluding the possibility of their transformation into notum cells (Fig. 2F). In wild-type haltere discs, Pdm1 is localized in the prospective capitellum region that shows very few Pdm-1 expressing cells in nub mutant discs. In wild-type leg discs, Pdm-1 is localized in rings, the most proximal corresponding to the coxa and the most distal corresponding to the basitarsus. In the antennal disc, we have detected the protein in the arista and in a ring corresponding to the second antennal segment. Pdm-1 protein pattern is unaffected in these discs in nub mutants (data not shown).
Cell Lineage.
The cell proliferation pattern of nub mutant discs was studied in clones of multiple wing hair (mwh). We have in addition generated M+ clones to detect gross abnormal parameters of cell proliferation in the wing disc and to study the clonal restriction borders in mutant wings. M+ cells have an advantage in cell proliferation with respect to M/+ cells of the rest of the wing (26). mwh clones were initiated at 60 h AEL and mwh M+ clones were initiated at different developmental times. These developmental times had to be corrected to correspond with normal development because nub larvae are delayed at hatching between 24 and 36 h compared with controls. Thus, as controls for the 60-h AEL clones in nub flies, we used clones initiated at 36 h AEL in wild-type wings. The parameters of these cell lineage experiments are summarized in Table 1. The frequency of clones in nub flies is lower and their size is smaller than in controls, indicating that at the time of the irradiation, there were fewer cells in a nub wing anlage and that nub cells grow less than in wild type (Table 1, experiments 1 and 2). The veins of the nub wings do not act as restriction borders as in wild-type wings (27). The M+ clones were classified as belonging to one of two major groups, depending on their size and restriction features, one group of early-induced clones (60–108 h AEL; Table 1, experiment 4) and the other of clones initiated after 120 h AEL (Table 1, experiment 5). The frequency of clones in both groups is lower than normal. They are larger in the A–P axis than in the proximal–distal axis, in contrast with the shape of the clones in a wild-type background, which are elongated along the proximal–distal axis (Fig. 3 A and B). The most surprising phenotype of the early induced clones is that they cross both the A–P and the D–V boundaries (Fig. 3 A and B). Some clones cover nearly the whole wing blade including pattern elements from both A and P compartments. However, the cell lineage carried out in Df(2L)Prl/+ mutant showed normal parameters of growth and clonal restrictions (Table 1, experiment 6). As seen in Table 1, we failed to observe clones crossing compartment boundaries in no-Minute flies.
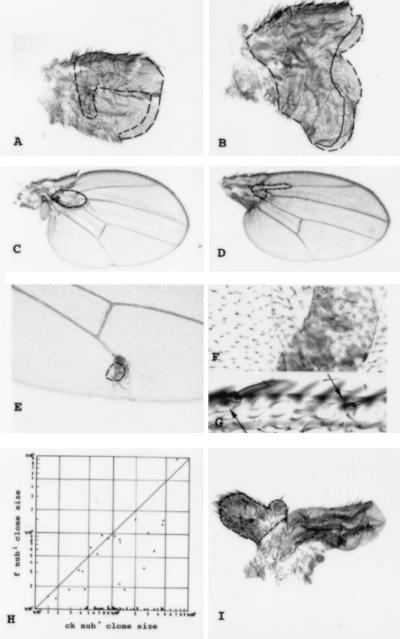
Figure 3.
Cell lineage and morphogenetic mosaics. (A and B) Different examples of early M+ clones (60–108 ± 12 h AEL) which do not respect the A–P and D–V restrictions (dashed line, contour of the dorsal component of the clone; solid line, ventral component). (C and D) nub mutant clones covering the proximal region of the wing blade. Note the small size of the clones (dashed lines, contour of the clone) and the reduction of the whole wing size. (D) Thickening of the proximal L2 is observed. (E and F) Tissue associated with f− cells but located between the two wing surfaces. The trichome pattern of this structure is characteristic of the hinge region (F is a magnification of E). (G) Bracteated bristles (bracts indicated by arrows) in a nub mutant clone covering the triple row. (H) Sizes of nub1 (mwh) clones and of their nub+ (f) twins in double logarithmic representation. (I) nub mutant wing containing a Tubα1>dpp clone (clone border is outlined in dashed lines).
Morphogenetic Mosaics.
Clones of nub cells were given the growth advantage of being associated with M+ in a nub M+/nub+ M phenotypically wild-type background. The parameters of this experiment are presented in Table 2 (experiments 1–3). nub mutant clones can appear anywhere in the wing blade. The phenotypes of nub1 mosaics are more extreme but otherwise similar to those of nub2. The frequency of nub clones is slightly reduced compared with controls, as is clone size, mainly for clones in the proximal region of the wing blade. The size of mutant cells is normal as indicated by the normal trichome density. The mutant clones differentiate ectopic bristles and campaniform sensilla, preferentially along L2 and L3. Clones including the veins differentiate in some cases veins thicker than those normally found in these positions, resembling the basal trunks of veins (Fig. 3D). Ventral clones covering the posterior crossvein fail to differentiate it. Some clones in intervein regions differentiate trichome densities and morphologies characteristic of those of more proximal axillar regions (may appear to correspond to the same level in the A–P axis; Fig. 3 E and F). Clones covering the triple row cause bracteated bristle differentiation (Fig. 3G). All of these pattern transformations can be understood as corresponding to shifts in cell differentiation of distal or medio–distal pattern elements to more proximal ones along the same A–P level of the wing. Proximal clones cause, in addition, nonautonomous effects on neighboring nonmutant cells causing a strong reduction in wing size. This size reduction affects not only the compartment that includes the mutant clone but also the opposite compartment (Fig. 3 C and D). We do not detect any folds in the surface of the wing opposite to the mutant clone surface, indicating that nonautonomous effects affect growth in both wing surfaces. This nonautonomous size reduction is independent of the size of the clone; in some cases the clone is formed by few cells.
Twin Clones.
We have done a twin-clone analysis to study the developmental parameters of nub mutant cells. The results (Table 2, experiments 4 and 5) show that nub cells proliferate less (by approximately 50%) than normal cells (Fig. 3H). Mutant cells do not have any preferential orientation in the proximal–distal axis with respect to the normal cells of the twin clone. We have detected normal clones without twin mutant clones (around 50%) anywhere on the wing disc. This effect is wing-specific because the frequency of normal clones without the mutant twin in tergites is comparable to controls (20% in nub twin experiment and 22% in controls), and the size of mutant clones is as in controls. The nub mutant differentiation phenotypes in veins, bristles and sensilla, and nonautonomous wing size reduction as a consequence of proximal mosaics were similar to phenotypes observed in nub clones induced in a M heterozygous mutant background.
Genetic Interactions.
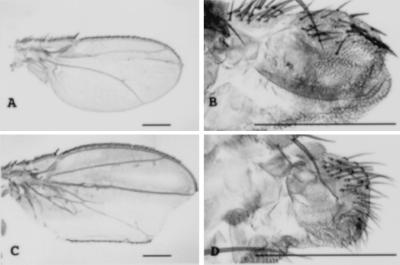
If a morphogenetic process is genetically perturbed in more than one step, a synergistic phenotype frequently results that is more extreme than the sum of the phenotypes of the individual mutants. We therefore have analyzed genetic combinations of nub and other mutations affecting genes with known morphogenetic functions. We have examined genetic combinations of nub and mutations in genes involved in cell communication (N, Dl, Ser) (28), in cell proliferation (top, ve, vn) (28), selector genes (en and ap) (28), and genes related to them (dpp, ptc, Cos, hh, Mrt, ciD) (28), in proximal–distal axis establishment (Dll, al) (14, 15, 29), in wing-specific development (vg) (30), and in wing shape (el, nw, cmp, sbd, wx, ll). All the phenotypes of the genetic combinations are additive (data not shown), with the exception of en, elbow (el) and dpp. The nub en double mutants are late embryonic lethals with cuticle phenotypes similar to those of pair-rule mutants (data not shown). This genetic interaction between en and nub could be related to nub function in embryos, a possibility that was not pursued further in this study. Although the phenotype of the dppd12/dppS8 heteroallelic combination by itself is a reduction in wing size and the absence of stretches of veins and the phenotype of el is a slight reduction in wing size and alteration of pattern of posterior veins, in the case of the wings of double mutant flies (nub el and dpp nub), only a few cells of the wing pouch are present but the hinge region remains unaffected (Fig. 4). We have studied clones of nub-el double mutant cells (Table 2, experiments 6–11). Although clonal analysis of el showed a slight reduction in clone size, the viability and size of the double mutant clones are severely reduced in both M+ and twin analysis with respect to the nub clones. Moreover el M+ clones show a severe transformation of distal structures of the wing into more proximal ones (data not shown). The combination of nub and a disk lethal allele of dpp in clones (Table 2, experiment 12) reveals very similar features to those of nub clones. To study further the genetic interaction between nub and dpp, we induced dpp ectopic expression clones in a nub mutant background. Clones of dpp-expressing cells (n = 21) cause some overgrowth but exclusively of dpp-expressing cells (Fig. 3I), in contrast to dpp ectopic expression clones in wild-type wings that causes overproliferation of the surrounding wild-type tissue in response to the ectopic expression in clones (7).
Figure 4.
Genetic interactions. (A) dppS8/dppd12 mutant wing. (B) nub dppS8/nub dppd12 mutant wing. (C) el1 mutant wing. (D) nub el1 mutant wing. The bars correspond to the same real size (B and D are 5× magnified).
Expression Patterns.
The absence of compartmentalization in adult nub mutant wings, observed in cell lineage M+ experiments, could be due to the misexpression of selector genes, and as a consequence, the expression of genes associated with the compartment boundaries, such as dpp, could be affected. We have monitored the expression of en and dpp from the beginning of the second larval instar to the end of larval development in nub mutant discs, and we have not found any difference from wild-type wings (Fig. 5 A, B, E, and H and data not shown). We have also studied wg (31) expression in nub mutant wings from early larval stages. The early expression pattern is normal and abnormalities of the expression pattern observed in third larval instar wing discs are due to the abnormal morphology of nub mutant wing discs (Fig. 5 C, D, and F). The expression patterns of ap (8) and vg (32) in mature nub mutant wing discs are normal (Fig. 5G and data not shown). Thus, the failures in compartmentalization cannot be attributed to misexpression of any of the studied genes.
Figure 5.
Expression patterns. (A and B) dpp expression pattern in mid second larval instar wild-type (A) and nub (B) wing disc. (C and D) wg expression pattern in mid second larval instar wild-type (C) and nub mutant (D) wing disc. (E) dpp expression pattern in third larval instar nub mutant wing disc. (F) wg expression pattern in third larval instar nub mutant wing disc. (A–F stainings were performed by using digoxigenine-labeled DNA probes). (G) ap expression pattern in nub mutant mature wing disc [anti-β-galactosidase staining using the aprk568 stock, which expresses lacZ in the same pattern as the ap RNA (8)]. (H) Anti-En antibody staining in third larval instar nub mutant wing disc.
DISCUSSION
The Pdm-1 protein has been proposed to be the putative protein encoded by the nub gene (16). The protein appears in the presumptive wing region of the wing disc from early second instar (150 cells) until pupation. Pdm-1 accumulates in the wing pouch region in third larval instar wing discs and is restricted to vein cells later and in pupae. The hinge region has no Pdm-1 protein, consistent with the absence of any alteration in this region in nub mutant flies. This region is associated with a late proximal–distal compartmentalization (1).
The phenotype of the nub wing is a reduction in wing cell number and alterations of wing patterns with a general proximalization of their elements. The cell lineage of nub mutant wings has shown that nub wings have fewer cells since the beginning of the larval development, in agreement with the idea that nub functions from the early disc proliferation period. We suggest that the decrease in wing cell number observed in nub mutant wings is due to a lower number of cell divisions caused by the absence of Pdm-1 protein during the entire proliferation period and not to cell death because we observed that cell death is restricted to the axillar wing region and only in third larval instar discs as in controls. The twin clone experiments confirm this idea as the mutant clones are around 50% smaller than control clones or fail to grow in 50% of the recombination events. Mutant cells fail to grow in all wing regions but not in other fly structures, such as tergites, indicating that the nub gene is specifically required in all wing blade cells. In morphogenetic mosaics, the cells of mutant nub clones show D–P transformations. Moreover mutant clones localized in the proximal region of the wing blade reduce growth of the whole wing irrespective of clone size, both in the surface containing the mutant cells and in the opposite wing surface, in both A and P compartments and in all wing axes.
We interpret nub reduced growth as the consequence of mutant cells having proximalized “positional values.” These positional values are scalar values corresponding to relative positions along the A–P, D–V, and proximal–distal axes, with an increasing maximum in compartment boundaries and at a minimum farthest away from these boundaries. The Entelechia model (28) proposes that proliferation results from the increase of values in the boundaries and from the intercalation of intermediate values between cells by cell division. Growth continues until the border values are maximal and the differences in the positional values between adjacent cells become minimal when the primordium reaches its species-specific size. The positional values reflect the amount of gene activity of so called “martial” genes. We suggest that the nub gene could be a martial gene, controlling the generation of positional values in the proximal–distal axis. In morphogenetic mosaics, the wild-type cells surrounding the nub mutant clone would receive signals from the mutant cells that are lower than normal for their position, as well as signals from their wild-type neighbors. The consequent intercalary growth results in a compromise between the positional values received, a process called “accommodation” (33) that affects the whole wing. Because, in the model, the positional value of a given cell results from the integration of all the scalar signals received from all its neighbors, the reduction in the positional value in the proximal–distal axis caused by nub mutation would affect the positional values of all cells and, hence, the proliferative behavior along all of the wing axes. Thus nub mosaics cause accommodation through both A–P and D–V compartment boundaries and, hence, growth of opposite compartments. This explanation is borne out by several observations. The shape of the M+ clones in nub wings perpendicular to the proximal–distal axis indicates failures in the generation of highest positional values along this axis (Fig. 6) resulting in an expansion in growth in the A–P axis. The surprising result that M+ clones induced in a nub mutant background respect neither the A–P nor the D–V compartment border could in principle be due to misexpression of selector genes and subsequent failures in compartmentalization. We have shown, however, that the selector genes are correctly expressed as are other genes relevant in disc growth and patterning. Thus, the clonal transgression of the compartment boundaries suggests that the lineage segregation could be a process genetically independent of selector gene expression maintenance. We propose that, in addition to the proximalization of nub mutant cells, these cells are prevented from acquiring high positional values characteristic of compartment boundaries, which in the model are maximal and responsible for the clonal restriction (Fig. 6). Thus, clonal restriction would be associated with scalar values and not only with the confrontation of territories expressing different selector genes. In nub mutant wings, with their phenotype enhanced by a M background, the positional values along the boundary of confrontation of cells expressing different selector genes would not be high enough to constitute a clonal restriction. The cells crossing from one compartment to another have to go through respecification of selector genes. This respecification could be carried out by cell–cell communication, in a process similar to that observed in the “apogenetic” mosaics in haltere mutants for hypomorphic bx alleles (34), where cells with haltere histotype (and Ubx protein expression) and cells with wing histotype (Ubx absence) can belong to the same clone. Morphogenetic mosaics of nub mutant cells in phenotypically normal surrounding tissue do respect the clonal restriction because in this situation, the normal cells surrounding the mutant clone reach the high positional values necessary to behave as a compartment border.
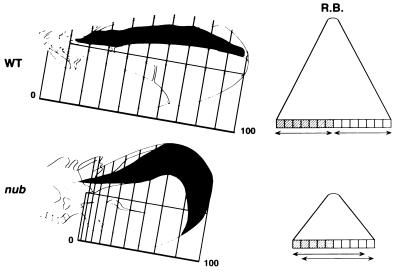
Figure 6.
Proximal–distal axis positional values and formation of compartment borders. In wild-type wings, the positional values along the proximal–distal axis (vertical bars) are correctly established by normal cells and the clones are elongated along the proximal–distal axis. Compartment boundaries (restiction boundaries = RB) in wild-type wings are the consequence of the high positional values reached by the cells in characteristic regions associated with the confrontation of cells expressing different selector genes (represented as squares with differential shading). The clones (horizontal arrows) never cross these compartment boundaries. In nub wings the proximal–distal positional values are not correctly established and nub cells are unable to reach the highest distal values, provoking the growth of the clones perpendicular to the A–P boundary. Clones in nub wings cross the compartment boundaries as a consequence of the inability of nub cells to meet the high positional values required for clonal restrictions.
The phenotypes observed in nub el double mutant flies and clones indicate a genetic interaction between these two genes. The transformations of distal regions into more proximal ones in nub el mutant clones are very strong and occur at a high frequency, suggesting that el could be acting with nub in the specification of values along the proximal–distal axis. nub dpp double mutant flies also have a synergistic phenotype. We have found that ectopic clonal dpp expression in nub wings does not give rise to overproliferation of the non-dpp-expressing cells as it does in a wild-type background (7). Thus, dpp could be one of the genes involved in generating the high positional values of the A–P compartment boundary, as signals for proliferation through intercalation of intermediate values away from the border; dpp-expressing cells, however, could not reach maximal values in a nub mutant background or nub cells cannot respond to it and proliferation would consequently not follow.
Table 1.
Cell lineage
| Exp. | Genotype | Age irrad. | No. clones (Freq.) | Clone size (No. cells) % total size | % A–P | % D–V |
|---|---|---|---|---|---|---|
| 1 | mwh/+ | 32–40 | 5 (6) | (700) | 0 | — |
| 2 | nub2; mwh/+ | 60 | 13 (2.7) | (310) | 0 | — |
| 3 | mwh/M | 60 | 32 (17.8) | 30 | 0 | 20 |
| 4 | nub2; mwh(h)/M | 60–108 | 33 (6.5) | 45 | 45.5 | 45.5 |
| 5 | nub2; mwh/M | 120 | 13 (16.2) | <10 | 0 | 0 |
| 6 | Df Prl/+; mwh/M | 60–70 | 10 (14.1) | 25 | 0 | 20 |
M used was M(3)67C. Ages ± 12 h AEL, except for experiment 1, which represents the exact time interval. Frequency (Freq.) is percent. M+ clone size is measured as the average of the percentage of the total wing size occupied by the clone. % A–P and % D–V represent the percentage of the clones that cross the A–P or D–V boundary, respectively.
Table 2.
Morphogenetic mosaics
| Exp. | Genotype | Age irrad. | No. clones* (Freq.) | Clone size, No. regions (mutant/twin)† | Ratio twin sizes |
|---|---|---|---|---|---|
| 1 | f; +/Mf+ | 72 | 31 (14) | 0.8 | |
| 2 | f; nub/Mf+ | 72 | 75 (9) | 0.4 | |
| 3 | f; nub2/Mf+ | 72 | 10 (10) | 0.78 | |
| 4 | f; nub/f+ck | 60 | 39/74 | (87/173) | 0.58 |
| 5 | f; nub2/f+ck | 60 | 11/26 | (114/151) | 0.56 |
| 6 | f; el1/Mf+ | 72 | 17 (10) | 0.4 | |
| 7 | f; el1/Mf+ | 60 | 11 (6.8) | 0.74 | |
| 8 | f; el1/f+ck | 72 | 42/51 | (45/48) | 1.06 |
| 9 | f; el1/f+ck | 60 | 19/23 | (114/113) | 1.04 |
| 10 | f; nub el1/Mf+ | 60 | 6 (2.7) | <0.1 | |
| 11 | f36a; nub el1/f+ck | 60/70 | 9/33 | (22.5/118) | 0.4 |
| 12 | f; dppdnub/Mf+ | 60 | 24 (4.4) | 0.65 |
f, f36a, dppd, dppd12; M, M(2L)24F. f+ is a f minigene inserted in 30B. Ages ±12 h AEL. M+ clone size is measured as the average number of intervein regions occupied by the clone.
No. of mutant clones/no. of twin control clones. Frequency is percent.
Average no. of mutant cells per clone compared with twin. Ratio of twin sizes is average ratio of mutant clone cell no./twin clone cell no.
Acknowledgments
We thank T. B. Kornberg, J. F. de Celis, S. Campuzano, I. Rodríguez, A. Baonza, and C. Extavour for constructive comments on the manuscript. We also thank K. Basler for providing fly stocks; S. Poole and S. Cohen for the anti-Pdm-1 and anti-Teashirt antibodies; P. Martín, A. López, and R. Hernández for technical assistance. F.J.C. is a postdoctoral fellow of the Fundación Rich. This work was supported by Grants PB92-0036 from the Dirección General de Investigaciín Científica y Técnica, SC1-CT92-0768 from the European Community and an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.
ABBREVIATIONS
- A
anterior
- P
posterior
- D
dorsal
- V
ventral
- AEL
after egg laying
References
- 1.García-Bellido A, Ripoll P, Morata G. Nat New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- 2.García-Bellido A. In: Results and Problems in Cell Differentiation. Ursprung H, Nöthiger R, editors; Ursprung H, Nöthiger R, editors. Vol. 5. Berlin: Springer; 1972. pp. 59–91. [Google Scholar]
- 3.García-Bellido A, Santamaría P. Genetics. 1972;72:87–101. doi: 10.1093/genetics/72.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillen I, Mullor J L, Capdevila J, Sánchez-Herrero E, Morata G, Guerrero I. Development (Cambridge, UK) 1995;121:3447–3456. doi: 10.1242/dev.121.10.3447. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg T, Siden I, O’Farrell P, Simon M. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- 6.Tabata T, Schwartz C, Gustavson E, Ali Z, Kornberg T B. Development (Cambridge, UK) 1995;121:3359–3369. doi: 10.1242/dev.121.10.3359. [DOI] [PubMed] [Google Scholar]
- 7.Zecca M, Basler K, Struhl G. Development (Cambridge, UK) 1995;121:2265–2278. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- 8.Cohen B, MacGuffin M E, Pfeifle C, Segal D, Cohen S M. Genes Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- 9.Tabata T, Kornberg T B. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 10.Posakony L G, Raftery L A, Gelbart W M. Mech Dev. 1991;33:69–82. doi: 10.1016/0925-4773(90)90136-a. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Irvine K D, Carroll S B. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- 12.Díaz-Benjumea F J, Cohen S M. Development (Cambridge, UK) 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- 13.Capdevila J, Guerrero I. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen S M, Jurgens G. EMBO J. 1989;8:2045–2055. doi: 10.1002/j.1460-2075.1989.tb03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen S M, Jurgens G. Roux’s Arch Dev Biol. 1989;198:157–169. doi: 10.1007/BF02438941. [DOI] [PubMed] [Google Scholar]
- 16.Ng M, Díaz-Benjumea F J, Cohen S M. Development (Cambridge, UK) 1995;121:589–599. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- 17.Billin A N, Cockerill K A, Poole S J. Mech Dev. 1991;34:75–84. doi: 10.1016/0925-4773(91)90045-8. [DOI] [PubMed] [Google Scholar]
- 18.Dick T, Yang X, Yeo S, Chia W. Proc Natl Acad Sci USA. 1991;88:7645–7649. doi: 10.1073/pnas.88.17.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd A, Sakonju S. Mech Dev. 1991;36:87–102. doi: 10.1016/0925-4773(91)90075-h. [DOI] [PubMed] [Google Scholar]
- 20.Lindsley D, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 21.Domínguez M, Campuzano S. EMBO J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spreij T E. Neth J Zool. 1971;21:221–261. [Google Scholar]
- 23.Cubas P, de Celis J F, Campuzano S, Modolell J. Genes Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- 24.Gómez-Skarmeta J L, Diez del Corral R, De la Calle-Mustienes E, Ferres-Marco D, Modolell J. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- 25.Ng M, Díaz-Benjumea F J, Vincent J P, Wu J, Cohen S M. Nature (London) 1996;381:316–318. doi: 10.1038/381316a0. [DOI] [PubMed] [Google Scholar]
- 26.Morata G, Ripoll P. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 27.González-Gaitán M, Capdevila M P, García-Bellido A. Mech Dev. 1994;46:183–200. doi: 10.1016/0925-4773(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 28.García-Bellido A, de Celis J F. Annu Rev Genet. 1992;26:275–302. doi: 10.1146/annurev.ge.26.120192.001425. [DOI] [PubMed] [Google Scholar]
- 29.Campbell G, Weaver T, Tomlinson A. Cell. 1993;74:1113–1123. doi: 10.1016/0092-8674(93)90732-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Sebring A, Esch J J, Kraus M E, Vorwerk K, Magee J, Carroll S B. Nature (London) 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- 31.Couso J-P, Bate M, Martínez-Arias A. Science. 1993;259:484–489. doi: 10.1126/science.8424170. [DOI] [PubMed] [Google Scholar]
- 32.Williams J A, Paddock S W, Carroll S B. Development (Cambridge, UK) 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- 33.García-Bellido A, Cortés F, Milán M. Proc Natl Acad Sci USA. 1994;91:10222–10226. doi: 10.1073/pnas.91.21.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botas J, Cabrera C V, Garcia-Bellido A. Roux’s Arch Dev Biol. 1988;197:424–434. doi: 10.1007/BF00398994. [DOI] [PubMed] [Google Scholar]