Abstract
Protein prenyltransferases catalyze the covalent attachment of isoprenoid lipids (farnesyl or geranylgeranyl) to a cysteine near the C terminus of their substrates. This study explored the specificity determinants for interactions between the farnesyltransferase of Saccharomyces cerevisiae and its protein substrates. A series of substitutions at amino acid 149 of the farnesyltransferase β-subunit were tested in combination with a series of substitutions at the C-terminal amino acid of CaaX protein substrates Ras2p and a-factor. Efficient prenylation was observed when oppositely charged amino acids were present at amino acid 149 of the yeast farnesyltransferase β-subunit and the C-terminal amino acid of the CaaX protein substrate, but not when like charges were present at these positions. This evidence for electrostatic interaction between amino acid 149 and the C-terminal amino acid of CaaX protein substrates leads to the prediction that the C-terminal amino acid of the protein substrate binds near amino acid 149 of the yeast farnesyltransferase β-subunit.
Biological activity of various proteins, including Ras, lamin B, and yeast a-factor mating pheromone, requires covalent attachment of a 15 carbon prenyl lipid (farnesyl) by protein farnesyltransferase. A related enzyme, protein geranylgeranyltransferase-I, transfers a 20 carbon prenyl lipid (geranylgeranyl) to numerous proteins, including the Ras-related Rho, Rac, and Cdc42 proteins. Both farnesyltransferase and geranylgeranyltransferase-I function as heterodimers; they share an α-subunit but have distinct β-subunits, which are only 33% identical. Both enzymes catalyze the attachment of a prenyl lipid to a cysteine four amino acids from the C terminus of the protein substrate. The preferred substrates of the mammalian and yeast farnesyltransferases have serine, methionine, cysteine, glutamine, or alanine in the C-terminal position (1–3) and are often referred to as CaaX proteins, where C is cysteine, a is usually an aliphatic amino acid, and X is the C-terminal amino acid. The preferred substrates of the mammalian and yeast geranylgeranyltransferase-I usually have leucine in the C-terminal position (1, 3–8) and are often referred to as CaaL proteins. Farnesyltransferase and geranylgeranyltransferase-I exhibit a degree of cross-specificity for both lipid (3, 9–11) and protein (3, 6, 7, 12–14) substrates.
The discovery that the Ras oncoprotein required farnesylation for function (15, 16) prompted intensive studies of farnesyltransferase, in large part because inhibitors of farnesyltransferase may prove to be effective for anti-cancer therapy (17). Despite the recent observation that both farnesyltransferase and geranylgeranyltransferase-I can efficiently prenylate K-rasB in vitro (14), and presumably in vivo, farnesyltransferase-specific inhibitors slow growth of K-rasB tumors in mice (18–24). Suprisingly, geranylgeranyltransferase-I-specific inhibitor (25) can also reverse the transformed phenotypes conferred by activated K-rasB. These promising observations suggest that reducing prenylation of Ras, without completely blocking it, may be sufficient to inhibit uncontrolled growth of cancer cells. Alternatively, additional proteins that depend on farnesylation or geranylgeranylation for function may be required for cancer cell growth. For example, RhoB prenylation appears to be required for Ras-mediated transformation (26, 27).
A more complete understanding of how farnesyltransferase and geranylgeranyltransferase-I bind their protein and lipid substrates will reveal the molecular basis of the substrate specificity and cross-specificity of these enzymes. Genetic (28–32) and biochemical (33–35) studies have provided evidence that both the α- and β-subunits are involved in protein substrate binding and that the active site is likely to be at the interface of the two subunits. The recently published structure of rat farnesyltransferase reveals a hydrophilic cleft at the junction of the α- and β-subunits, which has been proposed to bind the CaaX protein, and a hydrophobic cleft within the α-α barrel structure of the β-subunit, which has been proposed to bind the farnesyl diphosphate substrate (36). Previous studies had strongly implicated cysteine-299 of the β-subunit in both zinc binding and catalysis (31, 37, 38), and the structural data demonstrate that cysteine-299 coordinates with a zinc ion and resides at the junction of the hydrophobic and hydrophilic clefts (36). However, the currently available data do not provide a definitive view of protein or lipid substrate binding or the mechanism of catalysis.
We report here the identification of mutant forms of the yeast farnesyltransferase β-subunit (Ram1p) with altered protein substrate specificity. Our genetic evidence strongly suggests that the C-terminal amino acid of the protein substrate is in close proximity to amino acid 149 of the β-subunit when the protein substrate is productively bound to the farnesyltransferase. In conjunction with the rat farnesyltransferase structural data (36), our observations lead to a prediction of the general orientation for binding of the CaaX sequence to the farnesyltransferase.
MATERIALS AND METHODS
Plasmids.
A series of plasmids with substitutions at the last codon of the a-factor structural gene were constructed by inserting a PCR product (encoding the last 7 amino acids of the a-factor precursor and the 3′-flanking sequence) between the BamHI and EcoRI sites of YCpL-MFA1′, a CEN LEU2 plasmid that carries the MFA1 promoter and coding sequence for the first 29 amino acids of the a-factor precursor inserted as a 605 bp BglII–BamHI fragment into the BamHI site of YCplac111 (39). The BamHI site in the MFA1 gene was created by site-directed mutagenesis of nucleotide 90 from C to T (M. Ashby, unpublished work). The two oligonucleotides used in a PCR with Perkin–Elmer Taq polymerase were C-TGG-GAT-CCA-GCA-TGT-GTT-ATT-NNN-TAG-TTT-C, which contained a BamHI site (underlined) and a mixture of all four nucleotides at the last codon (NNN), and TCA-CTG-TAT-ACG-GAA-TTC-TCA-GC, which contained an EcoRI site (underlined) and was complementary to the 3′-flanking sequence of the MFA1 gene except for the C in bold.
A series of YCpU-RAS2val19 plasmids with substitutions at the last codon was constructed in pJR1056, a URA3 CEN plasmid containing RAS2val19-BMS, a mutated RAS2 gene with a valine codon in place of glycine codon 19 and with BamHI, MluI, and SphI sites in place of the 3′ end of the gene. The RAS2val19-BMS gene was constructed by site-directed mutagenesis (40) of the RAS2val19 gene (1.9 kb ClaI–HindIII fragment) in vector pRS306 (41), with a mutagenic oligonucleotide RAS2-SMB (5′-GCG-TTT-CTA-CAA-CTA-TTT-GCA-TGC-TAC-ACG-CGT-TAG-GAT-CCG-CTC-TTG-GAG-GC), which replaced 33 nucleotides of RAS2 (from 945 to 977) with CTAACGCGTGTAGCATGC. The SphI, MluI, and BamHI sequences in the RAS2-SMB oligonucleotide are underlined. pJR1044 was sequenced to confirm the presence of the RAS2val19-BMS gene.
To create a library of Ras2val19 variants with all possible substitutions at the C-terminal or X position of the CaaX sequence, oligonucleotides were inserted between the BamHI and SphI sites of pJR1056, a derivative of pRS316 (41) that lacks the polylinker BamHI site and has a 1.9 kb ClaI–HindIII fragment carrying RAS2val19-BMS (from pJR1044) inserted into the polylinker. The RAS2-CTA oligonucleotide (5′-CT-GGC-ATG-CTA-TTA-NNN-TAT-AAT-ACA-ACA-GCC-ACC-GGA-TCC-ATA) was annealed to the RAS2-BM primer (5′-TAT-GGA-TCC-GGT-GGC-TG) and incubated with Klenow fragment of Escherichia coli DNA polymerase. The resulting double-stranded oligonucleotide was cut with BamHI and SphI and ligated into pJR1056. Plasmids were sequenced with fluorescent dye-terminator technology (Applied Biosystems). Using the same strategy, the RAS2-C319S oligonucleotide (CT-GGC-ATG-CTA-TTA-ACT-TAT-AAT-AGA-ACA-GCC-ACC-GGA-TCC-ATA) was used to construct the RAS2val19-SIIS gene, which encodes a variant of Ras2val19 that cannot be prenylated due to the substitution of serine for cysteine-319 in the CaaX sequence.
YCpL-RAM1 was constructed by inserting a 3.1 kb BamHI–XbaI RAM1 fragment from pJR982 (12) into the polylinker of YCplac111, a LEU2 CEN plasmid (39). To create mutations in the RAM1 gene, oligonucleotide site-directed mutagenesis of the YCpL-RAM1 plasmid was performed with the Transformer Site-Directed Mutagenesis kit (CLONTECH). The oligonucleotide sequences were TCAGGAGGGCCCTTTGGTXXXGGTCCTG, where XXX indicates the position of codon 149, which differed among the five mutagenic oligonucleotides. Plasmids YCpL-RAM1G149A, YCpL-RAM1G149D, YCpL-RAM1G149E, YCpL-RAM1G149K, and YCpL-RAM1G149R have GCA, GAT, GAA, AAG, and AGA for codon 149, which encode alanine, aspartic acid, glutamic acid, lysine, and arginine, respectively. To aid in mutant identification, these oligonucleotides were designed with an nucleotide substitution that created an ApaI site (underlined) but did not alter the amino acid sequence. Mutant plasmids were sequenced to identify plasmids with the desired mutations at codon 149. One mutagenesis reaction also yielded a plasmid that had an ApaI site but retained the wild-type glycine codon 149; this plasmid was designated YCpL-RAM1-ApaI.
pAV5 has a blunt-ended XhoI–NdeI fragment from pAV2 (42), containing the entire YDJ1 gene and upstream region, inserted into YEp24 (Avrom Caplan, unpublished work).
Strains.
Strains used in these studies were constructed by standard genetic manipulations and are listed in Table 1.
Table 1.
Strains
| Strain | Genotype | Source or ref. |
|---|---|---|
| JRY3443 | mata MATα sst2-4 (ochre) trpl his3 ura3 can1 possibly cry1r | M. Ashby |
| JRY5312 | mataΔp hmlaΔp hmraΔp ade2 leu2 lys2 ura3 mfa1::hisG mfa2Δ::hisG sst2Δ GAL1-STE3::HIS3 | V. Boyartchuk (3) |
| JRY5388 | mataΔp hmlaΔp hmraΔp ade2 leu2 lys2 ura3 mfa1::hisG mfa2Δ::hisG sst2Δ GAL1-STE3::HIS3 ram1-102 | V. Boyartchuk (43) |
| JRY5389 | mataΔp hmlaΔp hmraΔp ade2 leu2 lys2 ura3 mfal1::hisG mfa2Δ::hisG sst2Δ GAL1-STE3::HIS3 ram1-103 | This study |
| JRY5390 | mata1 ade2 leu2 lys2 ura3 can1 mfa1::hisG mfa2Δ::hisG [pJR157 (MATα URA3)] | This study |
| JRY5391 | MATa ade2 his3 leu2 trp1 ura3 can1 ram1Δ::ADE2 [pJR1302 (RAM1 TRP1)] | This study |
| JRY5392 | mata1/MATa ade2/ade2 HIS3/his3 lys2/LYS2 TRP1/trp1 ura3/ura3 can1/can1 RAM1/ram1Δ::ADE2 | This study |
| mfal::hisG/MFA1 mfa2Δ::hisG/MFA2 [pJR157 (MATα URA3)] [pJR1302 (RAM1 TRP1)] | ||
| JRY5393 | MATa ade2 his3 leu2 trp1 ura3 can1 ram1Δ::ADE2 mfa1::hisG mfa2Δ::hisG | This study |
ram1Δ mfa1 mfa2 strain (JRY5393) was a segregant from diploid strain JRY5392, which was formed by mating a strain lacking both a-factor genes, MFA1 and MFA2 (JRY5390), with a strain deleted for the farnesyltransferase β-subunit gene, RAM1 (JRY5391). The RAM1 gene in JRY5391 was deleted by one-step gene replacement (44) with the ram1 deletion allele, ram1Δ::ADE2, which was constructed by replacing a 1,633 bp HpaI fragment, including the entire RAM1 gene, with a 2,241 bp BglII fragment of ADE2 from plasmid pASZ11 (45).
Halo Assay.
The relative levels of a-factor production by various MATa strains were evaluated by pheromone diffusion (halo) assay. Approximately 4 × 106 MATa cells were suspended in H2O and spotted onto a solid rich medium that had been spread with a lawn (≈2.5 × 106 cells) of MATα sst2 cells (JRY3443). After 2–3 days at 30°C, the relative amounts of a-factor produced by each MATa strain were evident from the size of the zone of growth inhibition (or halo) of MATα cells surrounding the MATa cells. The sst2 mutation facilitates measurement of a-factor production by making MATα cells more sensitive to a-factor-induced arrest (45). The size of the halo is proportional to the amount of a-factor exported from the MATa cells.
RESULTS
A genetic selection designed to isolate mutants defective in processing of the mating pheromone a-factor led to the discovery of two genes, AFC1 and RCE1, encoding proteins involved in proteolytic processing of prenylated proteins (43). Although the selection was intentionally biased toward the identification of genes encoding the CaaX proteases, it also provided the opportunity to find mutations that alter the protein substrate specificity of farnesyltransferase. Briefly, the selection identified mutations that decrease the ability of yeast cells to produce and export fully processed a-factor, particularly when the CaaX sequence at the C terminus of a-factor was changed from CVIA to CAMQ. A strain expressing a-factor–CAMQ as the only source of a-factor was mutagenized and 92 mutants that produced very little or no functional a-factor were isolated. After transformation with a wild-type a-factor gene, 24 of these 92 mutants produced a substantial amount of functional a-factor. Among the 24 mutants that were much more defective in the processing of a-factor–CAMQ than wild-type a-factor, mutants M1 and M10 were complemented by the wild-type RAM1 gene on a plasmid (Fig. 1). These results suggested that a ram1 mutation was responsible for the a-factor production defect in the M1 and M10 mutant strains.
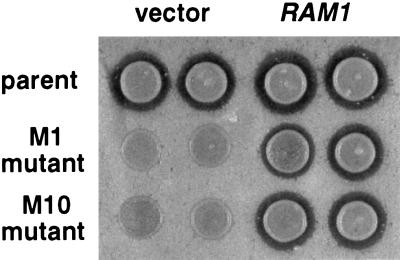
Figure 1.
RAM1 plasmid restored a-factor processing and export to the M1 and M10 mutants, as determined by a-factor halo assay. A CEN LEU2 RAM1 plasmid (YCpL-RAM1) or the CEN LEU2 vector (YCplac111) were transformed into the M1 and M10 mutant strains (JRY5388 and JRY5389) and the RAM1 parental strain (JRY5312 transformed with URA3 CEN MFA1-CAMQ plasmid, pJR1556). These MATa strains, which expressed a-factor–CAMQ as the only source of mating pheromone, were assayed for a-factor production by halo assay (see Materials and Methods). The indicated MATa strains were spotted onto a lawn of MATα sst2 cells and grown for 3 days; biologically active a-factor exported from the MATa strains arrested growth of the MATα sst2 cells, forming a zone of growth inhibition (halo) that is proportional to the amount of a-factor produced.
RAM1 Alleles That Affected Substrate Specificity of Farnesyltransferase.
To determine whether mutations were present in the RAM1 genes of the M1 and M10 mutant strains, the RAM1 genes from these two strains (JRY5388 and JRY5389) and from the wild-type parental strain (JRY5312) were amplified, with PCR, and sequenced. The sequence from the parental strain (JRY5312) was identical with GenBank entries for RAM1, whereas the M1 and M10 mutant strains each had a single nucleotide substitution of an A for G at nucleotide 446 in the ORF (Fig. 2). This mutation changed codon 149 from a glycine (GGA) to a glutamic acid (GAA) codon. Because M1 and M10 were both isolated from the same pool of mutagenized cells and have identical sequences, they are likely to be clonal.
Figure 2.
M1 and M10 mutants have mutations in codon 149 of the RAM1 gene. (A) Nucleotides 424–468 of the RAM1 gene from wild-type (JRY5312) and M1 and M10 mutant strains (JRY5388 and JRY5389). The sequences of the ram1–102 and ram1–103 alleles, in M1 and M10, respectively, were identical and differed from the wild-type RAM1 sequence in having an A at nucleotide 446 rather than a G, which changed codon 149 to GAA (a glutamate codon, underlined) from GGA (a glycine codon). (B) Predicted sequence of amino acids 135–172 of mutant Ram1p (M1 and M10 yeast FTβ), wild-type Ram1p (yeast FTβ; ref. 63 and this study), and homologous regions from human farnesyltransferase β-subunit (human FTβ; ref. 32) and tomato farnesyltransferase β-subunits (tomato FTβ; ref. 54). The G149E amino acid substitution in the ram1–102 and ram1–103 mutants (underlined) was in a highly conserved position in farnesyltransferase.
The level of Ram1p in the M1 and M10 mutant strains was indistinguishable from the level in the parental strain as determined by immunoblot (data not shown). Thus, the defect in a-factor production in the mutants could not be explained by an effect of the G149E substitution on the steady-state Ram1p concentration.
To determine whether or not the mutation that changed glycine-149 to glutamate (G149E) altered the specificity of the farnesyltransferase for its protein substrates, we assayed the in vivo substrate specificity of the wild-type and mutant farnesyltransferases. The ram1G149E strain (JRY5389) and the RAM1 parental strain (JRY5312) were transformed with mutated MFA1 plasmids encoding a-factor proteins with substitutions at the C-terminal (or X) position of the CaaX sequence. The relative levels of active a-factor produced from each of the altered MFA1 genes was determined by halo assay (Fig. 3). As expected, the RAM1 strain produced a large halo when a-factor terminated with the wild-type alanine (CVIA), or with cysteine (CVIC) or glutamine (CVIQ), which are the C-terminal amino acids of the known farnesyltransferase substrates, Ras1p (CIIC) and Ydj1p (CASQ), respectively. A large halo was also observed when a-factor terminated with glycine (CVIG). Small halos were observed when a-factor terminated with glutamate (CVIE), aspartate (CVID), tryptophan (CVIW), and lysine (CVIK); and virtually no halo was observed with a-factor terminating with arginine (CVIR). For seven of the nine a-factor variants (a-factor terminating with A, C, G, Q, E, D, or W), the halo produced by the ram1G149E strain was smaller than the halo produced by the RAM1 strain, which implied that the Ram1G149E protein could simply have a reduced effectiveness relative to the wild-type Ram1 protein. However, the differences in the relative halo sizes produced from a-factor–CVIW and a-factor–CVIG in the ram1G149E and RAM1 strains suggested that the G149E substitution altered the protein substrate specificity of the farnesyltransferase. Wild-type farnesyltransferase clearly produced a larger halo with CVIG than with CVIW, whereas ram1G149E–farnesyltransferase produced a larger halo with CVIW than with CVIG. Even stronger evidence for altered specificity was the ability of ram1G149E–farnesyltransferase to prenylate a-factor variants with a positively charged amino acid at the C terminus (CVIR and CVIK) and its inability to prenylate a-factor variants with a negatively charged amino acid at the C terminus (CVIE and CVID). With a-factor–CVIR, the halo produced by the ram1G149E strain was larger than the halo produced by the RAM1 strain, indicating that a-factor terminating with arginine could be farnesylated more effectively by ram1G149E–farnesyltransferase than by wild-type farnesyltransferase. With a-factor–CVIK, the halo sizes were equivalent in the ram1G149E and RAM1 strains, which implied that ram1G149E-farnesyltransferase and wild-type farnesyltransferase prenylate a-factor–CVIK equally well. However, additional experiments (described below) indicated that prenylation of Ste18p (the γ-subunit of the G protein involved in the mating pheromone response pathway) was decreased in the ram1G149E strain compared with the RAM1 strain, which would reduce a-factor induction by α-factor and consequently diminish the size of all a-factor halos produced by the ram1G149E strain compared with the RAM1 strain. Based on this additional information, we concluded that a-factor terminating with lysine was farnesylated more effectively by ram1G149E–farnesyltransferase than by wild-type farnesyltransferase.
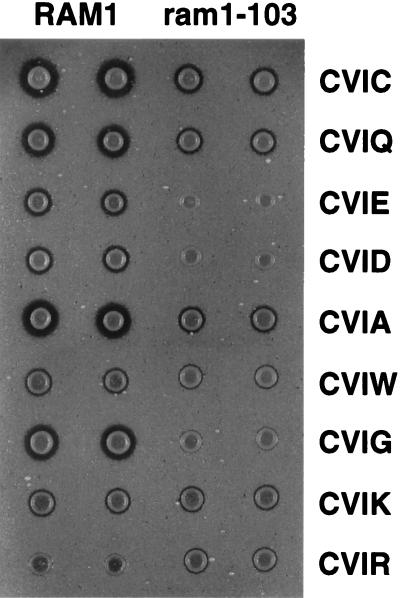
Figure 3.
Differential processing of a-factor variants by wild-type and mutant farnesyltransferases demonstrated that the ram1–103 mutation altered the protein substrate specificity of farnesyltransferase. The ram1–103 mutant strain (JRY5403) and the RAM1 parental strain (JRY5312) were transformed with mutated YCpL-MFA1 plasmids encoding a-factor variants, as indicated. Note that wild-type a-factor terminates with CVIA. The relative levels of a-factor produced by these strains was determined by halo assay (see Materials and Methods).
These halo assay results suggested that substitution of glutamate for glycine-149 of Ram1p affected the region of the farnesyltransferase responsible for interaction with the protein substrate. The positive charges at the C termini of a-factor–CVIR and a-factor–CVIK may be accommodated better by ram1G149E–farnesyltransferase because of a charge attraction between the positively charged arginine or lysine and the negatively charged glutamate 149. If this hypothesis were correct, then glycine-149 would be in, or very near, the protein substrate binding site of farnesyltransferase.
Interactions Between Farnesyltransferase Variants and Ras Protein Variants.
To test the hypothesis of direct interaction between amino acid 149 of Ram1p and the C-terminal amino acids of the CaaX substrate, mutant forms of RAM1 encoding Ram1pG149E, Ram1pG149D, Ram1pG149R, Ram1pG149K, or Ram1pG149A were created by site-directed mutagenesis of plasmid YCpL-RAM1. The ability of these mutant proteins to prenylate CaaX substrates with various C-terminal amino acid substitutions was assayed in vivo. To determine whether the interactions observed with a-factor substrates reflected a general feature of farnesyltransferase interaction with its substrates, variants of the CaaX protein Ras2val19 were used for these experiments. The Ras2val19 protein is similar to human Rasval12 protein; it is constitutively active due to a decreased ability to hydrolyze GTP. In yeast, constitutively active GTP-bound Ras2val19 protein results in several phenotypes, including low viability and heat-shock sensitivity under conditions of nutrient limitation. Because Ras2val19 dominant function requires prenylation (46, 47), the level of Ras2val19p prenylation is inversely related to the viability of the strains. A ram1Δ strain (JRY5393) was transformed with LEU2 CEN plasmids YCpL-RAM1-ApaI, YCpL-RAM1G149A, YCpL-RAM1G149D, YCpL-RAM1G149E, YCpL-RAM1G149R, YCpL-RAM1G149K, or vector YCplac111. The resulting strains were then transformed with altered RAS2val19 URA3 CEN plasmids encoding CaaX variants of Ras2val19p. In ram1Δ strains that coexpressed Ras2val19p (with the wild-type CaaX sequence CIIS) and either wild-type Ram1p or a mutant form of Ram1p (with G149D, G149E, G149R, G149K, or G149A substitutions), viability upon starvation was substantially lower than viability of a strain expressing Ras2val19p, but lacking Ram1p (Fig. 4A). These observations indicate that mutant forms of farnesyltransferase with substitutions of D, E, R, K, or A for Ram1p glycine-149 were able to prenylate and produce active Ras2val19p, when the C terminus of Ras2val19p was wild type (CIIS). In contrast, only the Ram1p variants with a negatively charged amino acid 149 (G149D and G149E) were able to effectively farnesylate Ras2val19p variants with positively charged C-terminal amino acids (CIIR and CIIK, Fig. 4 B and C). Moreover, the Ram1p variants with a positively charged amino acid 149 (G149R and G149K) were most effective in farnesylating Ras2val19p variants with the negatively charged aspartate (CIID) as the C-terminal amino acid, whereas the Ram1p variants with a negatively charged amino acid were the least effective (Fig. 4D). In strains expressing the nonprenylatable Ras2val19–SIIS protein in combination with any Ram1p variant, the viability upon starvation was similar to the viability of strains lacking Ram1p and expressing either Ras2val19–SIIS protein (Fig. 4E), Ras2val19–CIIS protein or any other Ras2val19p variant. Thus, the viability differences observed in Fig. 4 B–D were due to differences in the efficiency of Ras2val19p prenylation. The particular amino acid present at position 149 of Ram1p clearly influenced the ability of the enzyme to prenylate Ras2val19p variants with different amino acids at the C terminus. These data were most simply explained by a direct interaction between the charged amino acids at position 149 of Ram1p and the charged amino acids at the C terminus of Ras2val19p. Based on these data, we concluded that glycine-149 was very near the protein substrate recognition site of the farnesyltransferase.
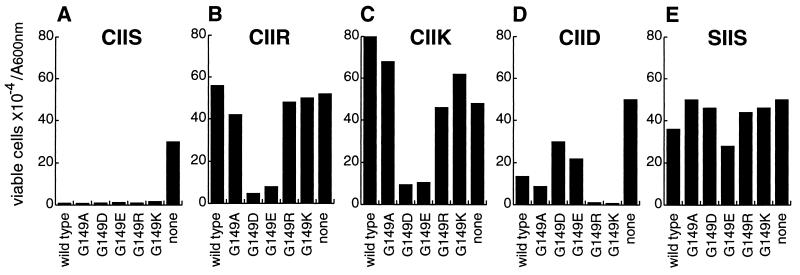
Figure 4.
Ram1p variants of farnesyltransferase exhibited allele-specific differences in the ability to prenylate Ras2val19 substrate variants in vivo. A ram1Δ strain (JRY5393) was transformed with plasmid YCpL-RAM1-ApaI (wild type), vector YCplac111 (none), or with a mutated YCpL-RAM1 plasmid that encoded a Ram1p variant with the indicated substitutions for glycine-149. The seven resulting strains were then transformed with plasmids, derived from pJR1056, that encode variants of Ras2val19 protein that differed in their CaaX sequences, as indicated. Wild-type Ras2val19 terminates with cysteine–isoleucine–isoleucine–serine (Ras2val19–CIIS; A). Variants that differed from wild type had the C-terminal serine substituted with arginine (Ras2val19–CIIR; B), lysine (Ras2val19–CIIK; C), or aspartate (Ras2val19–CIID; D). The Ras2val19–SIIS variant (E) could not be prenylated due to substitution of serine for cysteine-319, which is the prenyl lipid acceptor. To assess the extent of prenylation, viability of each strain following nutrient deprivation was determined following growth for 5 days at 25°C in liquid minimal medium lacking leucine and uracil (to select for the LEU2 and URA3 plasmids). Data are presented as viable cells × 10−4 per A600nm unit.
Ram1 Farnesyltransferase Variants Influence Prenylation of Other Substrates.
Because mutant forms of farnesyltransferase with substitutions at amino acid 149 have altered substrate specificity, they may fail to prenylate adequately one or more of the normal farnesyltransferase substrates in vivo. Indeed, ram1G149E–farnesyltransferase appears to be deficient in prenylation of Ste18p, the γ-subunit of the heterotrimeric G protein involved in the mating pheromone response pathway. MATa ram1-G149E cells were less sensitive to arrest by α-factor than MATa RAM1 cells (data not shown), strongly suggesting that ram1G149E–farnesyltransferase does not prenylate Ste18p well. In addition, our observation that a-factor halos produced by MATa ram1-G149E strains were generally smaller than the a-factor halos produced by MATa RAM1 strains (Fig. 1) is consistent with a Ste18p prenylation defect that results in decreased a-factor induction by α-factor. Moreover, halo assays revealed that an a-factor variant terminating with the C-terminal sequence of Ste18p (CTLM) was poorly prenylated by ram1G149E–farnesyltransferase compared with wild-type a-factor, whereas a-factor-CTLM and wild-type a-factor were prenylated to a similar extent by wild-type farnesyltransferase (data not shown). Together these data led us to conclude that ram1G149E–farnesyltransferase does not farnesylate Ste18p as well as wild-type farnesyltransferase.
Ram1p variants with basic or acidic amino acids at position 149 were not able to support growth at high temperatures, suggesting that they may be defective in prenylation of Ydj1p, a DnaJ homolog with CASQ at the C terminus that is required for growth at 37°C, but not at 30°C (48). Overproduction of Ydj1p from plasmid pAV5 (A. Caplan, unpublished work) partially suppressed the growth defect of ram1G149D, ram1G149E, and ram1G149R strains at 37°C. Moreover, a-factor–CVSA, which is similar to Ydj1p in having a serine at the a2 position of the CaaX sequence, was an adequate substrate for wild-type farnesyltransferase but not for ram1G149E–farnesyltransferase (data not shown).
DISCUSSION
The CaaX sequence is necessary and sufficient for substrate recognition by farnesyltransferase (2) and the identity of the C-terminal amino acid strongly influences substrate selection (1–3), but specific interactions between the protein substrate and the farnesyltransferase β-subunit have not been previously defined. We have presented strong genetic evidence for electrostatic interactions between Ram1p variants with charged amino acids at position 149 and CaaX substrate variants of a-factor and Ras2p with charged amino acids at the X position. Specifically, Ram1p, with a negatively charged amino acid substituted for glycine-149, was more competent than wild-type Ram1p in prenylation of CaaX substrates with a positively charged amino acid at the C terminus and was much less competent in prenylation of CaaX substrates with a negatively charged amino acid at the C terminus. Similarly, Ram1p, with a positively charged amino acid substituted for glycine-149, was more competent than wild-type Ram1p in prenylation of CaaX substrates with a negatively charged amino acid at the C terminus and was much less competent in prenylation of CaaX substrates with a positively charged amino acid at the C terminus. Therefore, our genetic data predict that Ram1p amino acid 149 is in very close proximity to the C-terminal amino acid of the CaaX sequence, providing evidence about the location of the protein substrate binding site of the farnesyltransferase β-subunit.
Glycine-149 is in a region of Ram1p that has high sequence similarity to other farnesyltransferase β-subunits (Fig. 2); a glycine is found in the analogous position of all published sequences [rat (50), bovine (51), human (32), pea (52), Arabidopsis (53), and tomato (54)]. The sequence surrounding glycine-149 lacks charged amino acids and includes a hydrophobic phenylalanine, which may explain why CaaX substrates with charged C-terminal amino acids are poor substrates for wild-type farnesyltransferase and why peptidomimetics with charged C-terminal amino acids (55) or a mispositioned carboxyl group (56) are poor inhibitors. The function of this conserved region has not been determined but it is part of the first of five internal repeated sequences present in farnesyltransferase β-subunits (57). The repeated sequences are 44–45 amino acids long and are comprised of a glycine-rich central region, which is an imperfectly conserved consensus sequence (GGFGGXPG), flanked by sequences that are somewhat less well conserved among the five repeats. The G in bold shows the position of G149 in repeat 1 of Ram1p. In rat farnesyltransferase (36), each repeated sequence in the β-subunit is comprised of two anti-parallel α-helices separated by the glycine-rich region. Some of the conserved amino acids in the β-subunit repeated sequences interact with conserved amino acids in the α-subunit repeated sequences (36), as originally proposed by Boguski et al. (57).
The prediction from our genetic data that glycine-149 is in close proximity to the C-terminal amino acid of CaaX substrates is consistent with the structure of the rat farnesyltransferase. The structure of rat farnesyltransferase reveals a hydrophilic cleft and a hydrophobic cleft, which have been proposed to bind the CaaX protein and the farnesyl diphosphate substrates, respectively (36). Cysteine-299 of the farnesyltransferase β-subunit, which is coordinated with a zinc ion (36) and has been proposed to be directly involved in catalysis (31, 37, 38), resides at the junction of these two clefts. The α carbon of glycine-142 of the rat farnesyltransferase β-subunit, which corresponds to glycine-149 of the yeast farnesyltransferase β-subunit, is located approximately 2 nm from the α carbon of cysteine-299. Therefore, if the cysteine of the CaaX sequence were near cysteine-299 and the CaaX peptide were in an extended conformation, the C terminus of the CaaX sequence could reside near glycine-142. Evidence from farnesyltransferase inhibitor studies (58, 59) is consistent with an extended conformation of the CaaX peptide, rather than the β turn conformation that had been previous proposed (18, 60).
Our prediction that the C-terminal amino acid of the CaaX substrate resides near glycine-142 of rat farnesyltransferase disagrees with the binding orientation of the CaaX substrate proposed by Park et al. (36). Their proposal is based on the supposition that the binding of the C-terminal tail of an adjacent β-subunit in the crystal lattice mimics some aspects of normal CaaX peptide binding. The binding of this tail may differ from the binding of CaaX substrates for a variety of reasons, including the fact that the sequence of the tail (proline–alanine–threonine–aspartate) differs significantly from the CaaX sequence and the fact that binding of the farnesyl diphosphate substrate, which is not present in the crystal, precedes binding of the CaaX substrate (61) and could affect enzyme or peptide substrate conformation. The genetic data presented here imply either that the binding of the C-terminal tail from an adjacent β-subunit in the crystal lattice does not accurately mimic the position of CaaX sequence binding or that the CaaX sequence binds farnesyltransferase in more than one position during the course of the farnesylation reaction.
Other amino acid substitutions that affect protein substrate recognition have been identified in farnesyltransferase β-subunits (29, 30, 32), but it is unclear whether or not the amino acids identified are directly involved in substrate binding. All of these amino acid substitutions are in a repeated sequence, either in the glycine-rich region or in the 10 amino acids following the glycine-rich region, which correspond to the inner α-helices of the α-α barrel. Interestingly, the sequences in the inner helices are the most highly homologous regions (67%–73%) among the farnesyltransferase β-subunits from different species. Mutations in the yeast geranylgeranyltransferase-I β-subunit gene that appear to affect protein substrate specificity (62) are also found in the repeated sequences.
In our studies, mutation of codon 149 resulted in farnesyltransferase variants that differentially affected prenylation and function of various CaaX proteins, including Ras2p, a-factor, Ste18p, and Ydj1p. Prenylation of Ras2val19p (CIIS) did not appear to be affected by substitutions at position 149 of Ram1p, whereas prenylation of Ste18p (CTLM) and Ydj1p (CASQ) by the Ram1p-G149E variant was reduced enough to partially or completely compromise the functions of these proteins. In general, the results obtained with a-factor and Ras2val19p variants terminating with a positively or negatively charged amino acid at the C terminus were similar, but there were some differences. The Ras2val19p variants terminating with arginine and lysine and a-factor terminating with arginine were prenylated very poorly by wild-type farnesyltransferase. However, a-factor terminating with lysine was prenylated to a significant extent. We propose that sequences outside the CaaX sequence of a-factor and Ras2val19p were responsible for differential interaction with farnesyltransferase. There is no sequence similarity between a-factor and Ras2p beyond the CaaX sequence and, although CaaX tetrapeptides can be prenylated in vitro by protein prenyltransferases, sequences outside the CaaX region can influence the prenylation efficiency (14).
Analysis of mutant enzymes has often led to insights on enzyme mechanisms. The data presented here provided a genetic test of a structure-based proposal for a protein substrate binding site and led to a different view of the orientation for binding of the CaaX substrate to the farnesyltransferase.
Acknowledgments
We thank Avrom Caplan for the YDJ1 plasmid (pAV5) and useful discussions, Shaul Yalovsky for valuable comments on the manuscript, and Marie Miles for help with yeast transformations. This work was supported by grants to J.R. from the California Tobacco Related Disease Research Program and from the National Institutes of Health (GM35827). Core support was provided by a grant from the National Institute of Environmental Health Sciences Mutagenesis Center.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Moores S L, Schaber M D, Mosser S D, Rands E, O’Hara M B, Garsky V M, Marshall M S, Pompliano D L, Gibbs J B. J Biol Chem. 1991;266:14603–14610. [PubMed] [Google Scholar]
- 2.Reiss Y, Stradley S J, Gierasch L M, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1991;88:732–736. doi: 10.1073/pnas.88.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caplin B E, Hettich L A, Marshall M S. Biochim Biophys Acta. 1994;1205:39–48. doi: 10.1016/0167-4838(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 4.Casey P J, Thissen J A, Moomaw J F. Proc Natl Acad Sci USA. 1991;88:8631–8635. doi: 10.1073/pnas.88.19.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinsella B T, Maltese W A. J Biol Chem. 1992;267:3940–3945. [PubMed] [Google Scholar]
- 6.Seabra M C, Reiss Y, Casey P J, Brown M S, Goldstein J L. Cell. 1991;65:429–434. doi: 10.1016/0092-8674(91)90460-g. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama K, Goodwin G W, Ghomashchi F, Glomset J A, Gelb M H. Proc Natl Acad Sci USA. 1991;88:5302–5306. doi: 10.1073/pnas.88.12.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finegold A A, Schafer W R, Rine J, Whiteway M, Tamanoi F. Science. 1990;249:165–169. doi: 10.1126/science.1695391. [DOI] [PubMed] [Google Scholar]
- 9.Pompliano D L, Schaber M D, Mosser S D, Omer C A, Shafer J A, Gibbs J B. Biochemistry. 1993;32:8341–8347. doi: 10.1021/bi00083a038. [DOI] [PubMed] [Google Scholar]
- 10.Adamson P, Marshall C J, Hall A, Tilbrook P A. J Biol Chem. 1992;267:20033–20038. [PubMed] [Google Scholar]
- 11.Armstrong S A, Hannah V C, Goldstein J L, Brown M S. J Biol Chem. 1995;270:7864–7868. doi: 10.1074/jbc.270.14.7864. [DOI] [PubMed] [Google Scholar]
- 12.Trueblood C E, Ohya Y, Rine J. Mol Cell Biol. 1993;13:4260–4275. doi: 10.1128/mcb.13.7.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohya Y, Qadota H, Anraku Y, Pringle J R, Botstein D. Mol Biol Cell. 1993;4:1017–1025. doi: 10.1091/mbc.4.10.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James G L, Goldstein J L, Brown M S. J Biol Chem. 1995;270:6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 15.Hancock J F, Magee A I, Childs J E, Marshall C J. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 16.Schafer W R, Kim R, Sterne R, Thorner J, Kim S H, Rine J. Science. 1989;245:379–385. doi: 10.1126/science.2569235. [DOI] [PubMed] [Google Scholar]
- 17.Sattler, I. & Tamanoi, F. (1996) in Regulation of the RAS Signaling Network, Molecular Biology Intelligence Unit Series, eds. Marula, H. & Burgess, A. W. (Landes, Austin, TX), pp. 95–137.
- 18.James G L, Goldstein J L, Brown M S, Rawson T E, Somers T C, McDowell R S, Crowley C W, Lucas B K, Levinson A D, Marsters J J. Science. 1993;260:1937–1942. doi: 10.1126/science.8316834. [DOI] [PubMed] [Google Scholar]
- 19.Kohl N E, Mosser S D, deSolms S J, Giuliani E A, Pompliano D L, Graham S L, Smith R L, Scolnick E M, Oliff A, Gibbs J B. Science. 1993;260:1934–1937. doi: 10.1126/science.8316833. [DOI] [PubMed] [Google Scholar]
- 20.Sepp L L, Ma Z, Rands E, Kohl N E, Gibbs J B, Oliff A, Rosen N. Cancer Res. 1995;55:5302–5309. [PubMed] [Google Scholar]
- 21.Kohl N E, Omer C A, Conner M W, Anthony N J, Davide J P, deSolms S J, Giuliani E A, Gomez R P, Graham S L, Hamilton K. Nat Med. 1995;1:792–797. doi: 10.1038/nm0895-792. [DOI] [PubMed] [Google Scholar]
- 22.Patel D V, Gordon E M, Schmidt R J, Weller H N, Young M G, Zahler R, Barbacid M, Carboni J M, Gullo B J, Hunihan L. J Med Chem. 1995;38:435–442. doi: 10.1021/jm00003a006. [DOI] [PubMed] [Google Scholar]
- 23.Manne V, Yan N, Carboni J M, Tuomari A V, Ricca C S, Brown J G, Andahazy M L, Schmidt R J, Patel D, Zahler R. Oncogene. 1995;10:1763–1779. [PubMed] [Google Scholar]
- 24.Leftheris K, Kline T, Vite G D, Cho Y H, Bhide R S, Patel D V, Patel M M, Schmidt R J, Weller H N, Andahazy M L, Carboni J M, Gullo B J, Lee F Y, Ricca C, Rose W C, Yan N, Barbacid M, Hunt J T, Meyers C A, Seizinger B R, Zahler R, Manne V. J Med Chem. 1996;39:224–236. doi: 10.1021/jm950642a. [DOI] [PubMed] [Google Scholar]
- 25.Lerner E C, Qian Y, Hamilton A D, Sebti S M. J Biol Chem. 1995;270:26770–26773. doi: 10.1074/jbc.270.45.26770. [DOI] [PubMed] [Google Scholar]
- 26.Prendergast G C, Khosravi F R, Solski P A, Kurzawa H, Lebowitz P F, Der C J. Oncogene. 1995;10:2289–2296. [PubMed] [Google Scholar]
- 27.Lebowitz P F, Davide J P, Prendergast G C. Mol Cell Biol. 1995;15:6613–6622. doi: 10.1128/mcb.15.12.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andres D A, Goldstein J L, Ho Y K, Brown M S. J Biol Chem. 1993;268:1383–1390. [PubMed] [Google Scholar]
- 29.Mitsuzawa H, Esson K, Tamanoi F. Proc Natl Acad Sci USA. 1995;92:1704–1708. doi: 10.1073/pnas.92.5.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Villar K, Mitsuzawa H, Yang W, Sattler I, Tamanoi F. J Biol Chem. 1997;272:680–687. doi: 10.1074/jbc.272.1.680. [DOI] [PubMed] [Google Scholar]
- 31.Fu H W, Moomaw J F, Moomaw C R, Casey P J. J Biol Chem. 1996;271:28541–28548. doi: 10.1074/jbc.271.45.28541. [DOI] [PubMed] [Google Scholar]
- 32.Omer C A, Kral A M, Diehl R E, Prendergast G C, Powers S, Allen C M, Gibbs J B, Kohl N E. Biochemistry. 1993;32:5167–5176. doi: 10.1021/bi00070a028. [DOI] [PubMed] [Google Scholar]
- 33.Reiss Y, Seabra M C, Armstrong S A, Slaughter C A, Goldstein J L, Brown M S. J Biol Chem. 1991;266:10672–10677. [PubMed] [Google Scholar]
- 34.Ying W, Sepp L L, Cai K, Aloise P, Coleman P S. J Biol Chem. 1994;269:470–477. [PubMed] [Google Scholar]
- 35.Pellicena P, Scholten J D, Zimmerman K, Creswell M, Huang C C, Miller W T. Biochemistry. 1996;35:13494–13500. doi: 10.1021/bi961336h. [DOI] [PubMed] [Google Scholar]
- 36.Park H W, Boduluri S R, Moomaw J F, Casey P J, Beese L S. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F L, Fu H W, Casey P J, Bishop W R. Biochemistry. 1996;35:8166–8171. doi: 10.1021/bi960574+. [DOI] [PubMed] [Google Scholar]
- 38.Huang C C, Casey P J, Fierke C A. J Biol Chem. 1997;272:20–23. doi: 10.1074/jbc.272.1.20. [DOI] [PubMed] [Google Scholar]
- 39.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 40.Zoller M J, Smith M. Nucleic Acids Res. 1982;10:6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caplan A J, Douglas M G. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyartchuk V L, Ashby M N, Rine J. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 44.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 45.Stotz A, Linder P. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 46.Powers S, Michaelis S, Broek D, Santa A S, Field J, Herskowitz I, Wigler M. Cell. 1986;47:413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- 47.Schafer W R, Trueblood C E, Yang C C, Mayer M P, Rosenberg S, Poulter C D, Kim S H, Rine J. Science. 1990;249:1133–1139. doi: 10.1126/science.2204115. [DOI] [PubMed] [Google Scholar]
- 48.Caplan A J, Tsai J, Casey P J, Douglas M G. J Biol Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- 49.Cyr D M. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- 50.Chen W J, Andres D A, Goldstein J L, Brown M S. Proc Natl Acad Sci USA. 1991;88:11368–113672. doi: 10.1073/pnas.88.24.11368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohl N E, Diehl R E, Schaber M D, Rands E, Soderman D D, He B, Moores S L, Pompliano D L, Ferro N S, Powers S, Thomas K A, Gibbs J B. J Biol Chem. 1991;266:18884–18888. [PubMed] [Google Scholar]
- 52.Yang Z, Cramer C L, Watson J C. Plant Physiol. 1993;101:667–674. doi: 10.1104/pp.101.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- 54.Yalovsky S, Trueblood C E, Callan K L, Narita J O, Jenkins S M, Rine J, Gruissem W. Mol Cell Biol. 1997;17:1986–1994. doi: 10.1128/mcb.17.4.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nigam M, Seong C M, Qian Y, Hamilton A D, Sebti S M. J Biol Chem. 1993;268:20695–20698. [PubMed] [Google Scholar]
- 56.Qian Y, Vogt A, Sebti S M, Hamilton A D. J Med Chem. 1996;39:217–223. doi: 10.1021/jm950414g. [DOI] [PubMed] [Google Scholar]
- 57.Boguski M S, Murray A W, Powers S. New Biol. 1992;4:408–411. [PubMed] [Google Scholar]
- 58.Qian Y, Blaskovich M A, Saleem M, Seong C M, Wathen S P, Hamilton A D, Sebti S M. J Biol Chem. 1994;269:12410–12413. [PubMed] [Google Scholar]
- 59.Vogt A, Qian Y, Blaskovich M A, Fossum R D, Hamilton A D, Sebti S M. J Biol Chem. 1995;270:660–664. doi: 10.1074/jbc.270.2.660. [DOI] [PubMed] [Google Scholar]
- 60.Stradley S J, Rizo J, Gierasch L M. Biochemistry. 1993;32:12586–12590. doi: 10.1021/bi00210a006. [DOI] [PubMed] [Google Scholar]
- 61.Furfine E S, Leban J J, Landavazo A, Moomaw J F, Casey P J. Biochemistry. 1995;34:6857–6862. doi: 10.1021/bi00020a032. [DOI] [PubMed] [Google Scholar]
- 62.Ohya Y, Caplin B E, Qadota H, Tibbetts M F, Anraku Y, Pringle J R, Marshall M S. Mol Gen Genet. 1996;252:1–10. [PubMed] [Google Scholar]
- 63.Goodman L E, Perou C M, Fujiyama A, Tamanoi F. Yeast. 1988;4:271–281. doi: 10.1002/yea.320040405. [DOI] [PubMed] [Google Scholar]