Abstract
Autonomously replicating sequence (ARS) elements, which function as the cis-acting chromosomal replicators in the yeast Saccharomyces cerevisiae, depend upon an essential copy of the 11-bp ARS consensus sequence (ACS) for activity. Analysis of the chromosome III replicator ARS309 unexpectedly revealed that its essential ACS differs from the canonical ACS at two positions. One of the changes observed in ARS309 inactivates other ARS elements. This atypical ACS binds the origin recognition complex efficiently and is required for chromosomal replication origin activity. Comparison of the essential ACS of ARS309 with the essential regions of other ARS elements revealed an expanded 17-bp conserved sequence that efficiently predicts the essential core of ARS elements.
The replication of yeast chromosomes is accomplished by the activation of multiple cis-acting replicators (for review, see ref. 1). These replicators were first identified by their ability to promote high-frequency transformation and stable extrachromosomal maintenance of plasmids in yeast (2, 3). On the basis of these properties, they were named autonomously replicating sequence (ARS) elements. Two-dimensional gel electrophoresis techniques for identifying replication intermediates were used to demonstrate that ARS elements colocalize with plasmid replication origins (4, 5). The analysis of chromosomal replication initiation by these techniques revealed that many ARS elements are cis-acting replicators required for chromosomal replication origin activity (6–9). However, only a subset of ARS elements are active as origins in their native chromosomal contexts (10, 11).
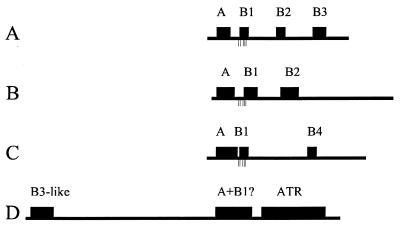
ARS elements are modular (Fig. 1). All contain an essential match or near match to the 11-bp ARS consensus sequence (ACS) WTTTAYRTTTW (where W is A or T, Y is T or C, and R is A or G; refs. 17 and 18). Mutations in this sequence abolish ARS function (for review, see ref. 1). ARS elements also contain additional near matches to the ACS that are dispensable in the ARS elements that have been examined (12–16). The mutation of several bases on either side of the essential match to the ACS also impairs ARS function; this larger element has been called domain A (12–16, 19, 20). The essential match to the ACS is recognized and bound by a six-subunit complex, the origin recognition complex (ORC), that functions as a DNA-dependent ATPase and is thought to be the replication initiator protein (21–26).
Figure 1.
Modular nature of ARS elements. The functional elements defined by mutational analysis of four ARS elements are indicated by boxes on the line diagrams. All four are aligned by their essential matches to the ACS and drawn to the same scale. The tick marks below the lines indicate the positions +8, +10, +15, +17, and +18 relative to the end of the essential match to the ACS (see text). (A) ARS1 (12). (B) ARS307 (13, 14). (C) ARS305 (15). (D) ARS121 (16). The region labeled A+B1? corresponds to the “core” element of Walker et al. (16). ATR is the A+T-rich domain. B3-like corresponds to the pair of ARS binding factor 1 binding sites.
In addition to domain A, a region 3′ to the T-rich strand of the ACS is also required; it is referred to as domain B (19). There appears to be little or no sequence conservation among ARS elements in the domain B region. Instead, domain B is composed of several small nonredundant sequence elements (B1 to B4) that can differ from one ARS to another (12–15). Despite a lack of sequence homology, some of these elements act as functional analogues. Thus, the B1 and B2 elements of ARS307 can substitute for their counterparts in ARS1, and vice versa (13). Some mutations in the B1 elements of ARS1 and ARS307 reduce the efficiency of ORC binding, suggesting that B1 functions along with domain A as part of the ORC binding site (27, 28). However, the observation that some B1 mutations reduce ARS efficiency without affecting ORC binding in vitro suggests that the B1 element plays an additional role in replicator activity (28). The B3 element of ARS1 is a binding site for ARS binding factor 1, a transcriptional activator and repressor. ARS binding factor 1 binding sites are found 5′ of the T-rich strand of the essential match to the ACS of ARS121 (Fig. 1D); these sites stimulate ARS activity from this position or when moved into domain B (16).
Despite our ability to broadly describe ARS structure, many specific issues are not resolved. In this manuscript we describe the analysis of ARS309, an active chromosomal replicator on the right arm of chromosome III. Detailed mutational analysis of the matches to the ACS within ARS309 led to the unexpected discovery that the essential ACS of ARS309 contains a mismatch in a group of three thymines that are invariant in the other 19 essential ACS elements defined to date. We show that this variant ACS is essential for chromosomal replicator activity and that it binds ORC in vitro. Comparison of this newly defined essential region of ARS309 with other ARS elements allowed the definition of an expanded ACS that substantially improves predictions of the essential ACS of other ARS-containing DNA fragments.
MATERIALS AND METHODS
Plasmids and Strains.
Plasmids ARS309–306 and ARS309–326 were made by cloning the ARS309-containing 0.35-kb EcoRI–BamHI fragment of pVH53 (29) in pRS306 (30) and pRS326 (14), respectively. The ARS309 fragment was derived from a genomic 0.35-kb EcoRI–HpaI fragment (chromosome III coordinates 131, 585 to 131, 933; MIPS-Yeast Genome Project, Sept. 2, 1996, release) by adding a BamHI linker to the HpaI site. The plasmid 309-flk was made by ligating the 1.4-kb HindIII–EcoRI fragment of J11D-5a (31), the 0.9-kb EcoRI–BglII fragment of J11D-2 (31), and pRS306 digested with HindIII plus BamHI. 309-flk was digested with HpaI, and a BamHI linker was inserted to create 309-flk+Bam.
Escherichia coli DH5α (Life Technologies, Grand Island, NY) was used for the propagation of plasmids. The diploid yeast strain 1C6 (14) was used for testing ARS activity, and the haploid yeast strain YP45 (30) was used for the analysis of chromosomal DNA replication intermediates.
Mutagenesis of ARS309.
Exonuclease III deletions of ARS309 were constructed as described (14). For deletions from the EcoRI end, ARS309–306 was digested with EcoRI plus KpnI; for deletions from the HpaI end, ARS309–326 was digested with BamHI plus SacI. The deletion fragment “57 to 135” is the genomic RsaI fragment cloned into pRS326 at the EcoRI site, which was blunted by filling in with T4 polymerase. The site-specific mutations in the matches to the ACS were generated by the method of Kunkel et al. (32), as described by Theis and Newlon (14). The oligonucleotides used were 22 nucleotides long and were designed to change two of the three thymidines at positions 8, 9, and 10 of the matches to the ACS to other bases. The alterations made are shown in Table 1. The plasmid ARS309–326 was used as the template for all the single mutations. The double and triple mutants were constructed by using the appropriate singly or doubly mutant plasmid as template with the desired oligonucleotide. Oligonucleotides were synthesized by the New Jersey Medical School Molecular Resource Facility.
Table 1.
ACS matches and mutations
| ACS match | Location, bp | Mutant version | Positions altered |
|---|---|---|---|
| ATCTATATTTT | 31–41 | ATCTATATaaT | 39, 40 T → A |
| TTATATGTTTA | 169–159 | TTATATGaaTA | 161, 162 T → A |
| TTTTATATGTT | 171–161 | TTTTATATGaa | 161, 162 T → A |
| TTTTTTATTTT | 241–231 | TTTTTTAaaTT | 233, 234 A → T |
| TTTGAAGTTTT | 75–85 | TTTGAAGTaaT | 83, 84 T → A |
| TTAAATGTTTT | 104–94 | TTAAATGTggT | 95, 96 A → C |
| GTTTATATCTT | 62–72 | GTTTATAgCgT | 69, 71 T → G |
Matches to the ACS in the 350-bp HpaI–EcoRI fragment. Positions that differ from the ACS are underlined. Alterations in the mutant versions are indicated as lowercase type. All matches are shown as the T-rich strand.
To integrate mutant ARS309 derivatives into the chromosome, the 0.35-kb EcoRI–BamHI fragment from the desired mutant plasmid was swapped for the wild-type 0.35-kb fragment in 309-flk+Bam. A two-step gene replacement (33) was performed in YP45 (30), using the unique EcoRI or BlpI sites to direct integration. Mutant popouts were identified by the presence of a new BamHI site detected by Southern blotting and/or PCR amplification of genomic DNA. PCR products amplified from the mutant strains were sequenced to confirm the presence of the mutations.
Examination of DNA Replication Intermediates.
Preparation of genomic DNA and two-dimensional gel electrophoresis were as described (14).
ORC Footprinting.
Probes were made by PCR as described (34) by using T7 primer (Life Technologies) end-labeled by incubation with polynucleotide kinase and [γ-32P]ATP and unlabeled M13 reverse primer (New England Biolabs). The plasmid templates for PCR were ARS309–326 and the appropriate mutant derivatives. Footprinting was performed as described by Klemm et al. (22). Recombinant ORC was the generous gift of Stephen Bell (Massachusetts Institute of Technology). Chemical sequencing reactions (A+G and T) were performed as described by Richterich et al. (35).
RESULTS AND DISCUSSION
ARS309 is associated with a chromosomal replication origin on the right arm of Saccharomyces cerevisiae chromosome III (36). The DNA sequences required for maximum plasmid stability are contained within a 0.35-kb EcoRI–HpaI genomic DNA fragment (29, 31). In contrast to ARS1 and ARS307 (12–14), ARS309 contains no perfect match to the ACS but rather contains four 10 of 11 matches (Table 1).
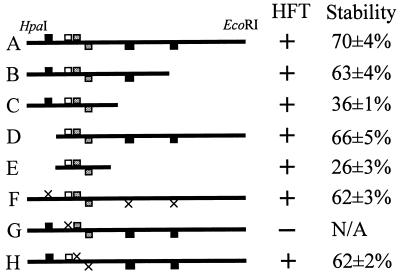
As a first step in localizing ARS309 within the 0.35-kb genomic DNA fragment, we constructed exonuclease III deletion series from either the EcoRI or HpaI sides. As shown in Fig. 2, deletions from the EcoRI side cause a gradual reduction in plasmid stability (constructs A, B, and C and data not shown). The small fragment containing nucleotides 1–147 (Fig. 2C) was still Ars+ despite deletion of three of the four 10 of 11 matches to the ACS. Deletion of 53 bp from the HpaI side had no effect on ARS function (Fig. 2D), despite removing the 10 of 11 match to the ACS at positions 31–41. The data from the two sets of unidirectional deletions suggest that all of the 10 of 11 matches are dispensable. This was confirmed in two ways. (i) Two-base changes were made in each 10 of 11 match, both singly and in combination; similar alterations of the ACS have been shown to abolish activity of other ARS elements (15, 18, 23, 37, 38). None of the single mutations had a significant effect on ARS function, nor did the triple mutation that altered all four 10 of 11 matches (Fig. 2F, Table 1, and data not shown). It was possible to alter all four 10 of 11 matches with three mutations because two of them overlap. (ii) An RsaI fragment retained ARS activity (Fig. 2E).
Figure 2.
Effect of deletions and point mutations on ARS309 function. Lines represent the sequences present in plasmids. Position 1 is the first adenine of the HpaI site; position 349 is the cytidine of the EcoRI site. Solid boxes show the 10 of 11 matches to the ACS at positions 31–41, 159–169, 161–171, and 231–241. The open box marks the position of the 9 of 11 match at positions 62–72. The stippled boxes show the positions of the 9 of 11 matches at positions 75–85 and 94–104. × indicates point mutations altering the ACS matches (see Table 1). Boxes above the line indicate that the T-rich strand of the ACS is in the top strand, and boxes below the line indicate the T-rich strand of the ACS is in the lower strand. HFT: +, high-frequency transformation; −, no high-frequency transformation. Stability: Numbers show percentage of plasmid bearing cells in a logarithmic-phase culture under selection (mean ± SEM). (A) Full-length wild-type ARS309 (base pairs 1–349). (B) Deletion mutant containing base pairs 1–227. (C) Deletion mutant containing base pairs 1–147. (D) Deletion mutant containing base pairs 54–349. (E) RsaI subclone (base pairs 57–135). (F) Triple point mutation altering all four 10 of 11 matches (see Table 1). (G) Mutation altering the 9 of 11 match at positions 62–72 (see Table 1). (H) Mutation altering the two 9 of 11 matches at positions 75–85 and 94–104 (see Table 1).
This 79-bp RsaI fragment contains three 9 of 11 matches to the ACS (Table 1). To determine whether one of these matches functions as the essential match to the ACS, 2-bp changes that alter each of these 9 of 11 matches were made in the 0.35-kb HpaI–EcoRI fragment. Mutation of the matches at positions 75–85 and 94–104 had no effect on ARS function, either singly or in combination (Fig. 2H). In contrast, ARS activity was abolished when the match at positions 62–72 was altered, identifying this as the essential match (Fig. 2G).
It was surprising to find that this 9 of 11 match, gTTTATATcTT, was the essential ACS. In particular, the TcT instead of TTT was surprising because all of the previously identified essential matches to the ACS conserved the three thymines at positions 8, 9, and 10 (39). In addition, any base-pair substitution at any one of the TTT sites inactivates ARS307 (18). Some changes at these positions have also been shown to inactivate ARS1, and these changes reduced ORC binding to ARS1 (23). ARS309 differs from all other ARS elements examined to date in that its essential match has a cytidine at position 9.
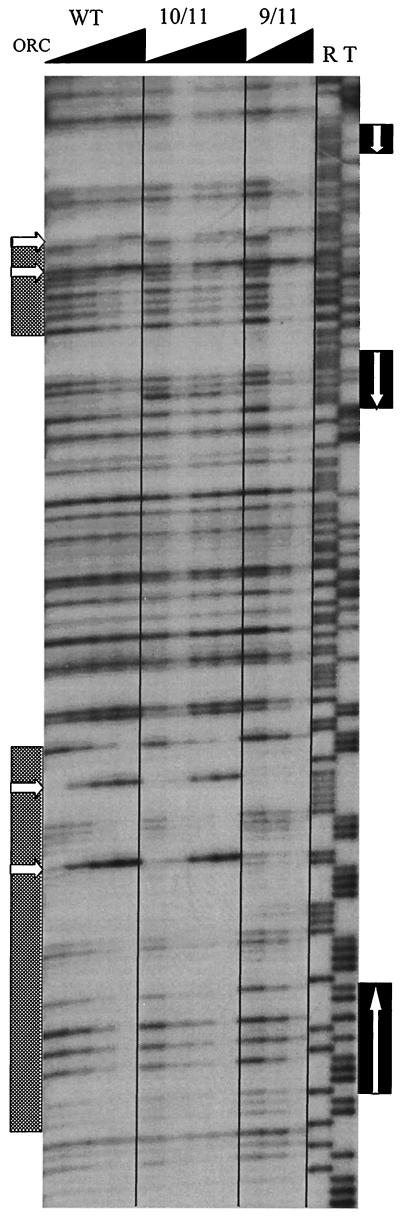
Because all essential matches to the ACS tested to date bind ORC, it was important to determine whether ORC binds to the exceptional ACS in ARS309. In other ARS elements, ORC protects a region of ∼50 bp that includes the essential ACS. ORC binding also induces DNase I-hypersensitive sites within the B domain, the most prominent hypersensitive sites occurring 12–16 and 22–26 bp away from the ACS. In vitro footprinting with purified ORC was performed on the 0.35-kb ARS309 fragment. As can be seen in Fig. 3, ORC protects the region from positions 60 to 102, which contains the essential ACS. Moreover, ORC binding induces two hypersensitive sites at positions 87 and 97, which are 15 and 25 bp away, respectively, from the ACS. The hypersensitive sites are detected in the wild-type fragment with as little as 5 ng of ORC, although 25 ng are required for full protection of this region. When the mutated ACS that abolishes origin function was tested for ORC binding, protection of the 60 to 102 region was abolished, even at 100 ng of ORC, and no hypersensitive sites were seen. The absence of hypersensitive sites at 100 ng of ORC indicates that the 2-bp mutation in the essential ACS reduced ORC binding at least 20-fold.
Figure 3.
ORC footprinting of ARS309 and mutant derivatives. End-labeled fragments were footprinted with ORC. WT, wild-type ARS309 (Fig. 2A); 10/11, ARS309 with all four 10 of 11 matches mutated (Fig. 2F); 9/11, ARS309 with the essential 9 of 11 match mutated (Fig. 2G). For WT and 10/11, the four footprinting reactions contained 0, 5, 12.5, and 25 ng of ORC; for 9/11, the three reactions contained 0, 25, and 100 ng. Lanes R and T are chemical sequencing reactions for A+G, and T, respectively. The hatched boxes to the left indicate the protected regions (positions 60–102, lower box; 180–204, upper box), and the arrows point to hypersensitive sites (positions 87, 97, 196/197, 205/206 from bottom to top). Solid boxes to the right of the sequencing reactions mark the positions of the 10 of 11 matches to the ACS at positions 231–241 (upper box), 159–169 and 161–171 (middle box), and the essential 9 of 11 match at positions 62–72 (lower box). Arrows within these boxes indicate the orientation of the T-rich strand of the ACS. Note that neither the protected regions nor the hypersensitive sites are altered in the 10 of 11 match mutant, but the 9 of 11 match mutant abolishes the protection and hypersensitive sites in the region from positions 60 to 102.
ORC protected neither the 10 of 11 match at positions 31–41 (data not shown) nor the two overlapping sites at positions 159–171. Although there was weak protection of the DNase I cleavage sites at positions 159, 165, and 167, there was no other protection in the region from positions ∼120 to 171, as would be expected for ORC binding at positions 159–171. In addition, there are no hypersensitive sites induced within this region. Because there are no DNase I cleavage sites in naked DNA in the region containing the fourth 10 of 11 match (positions 231–241), we cannot assess protection by ORC in this region. However, binding of ORC to this match to the ACS would be expected to protect the region from positions ∼190 to 241, and it is clear that the DNase I cleavage sites at positions 217 and 221 were not protected. Furthermore, the absence of hypersensitive sites within this region suggests that ORC does not bind to this match. We also examined ORC binding to the fragment containing mutations in all four 10 of 11 matches to the ACS. As seen with the wild-type fragment, there is protection of the region from positions 60 to 102 and induction of the two hypersensitive sites at positions 87 and 97. The amount of ORC required for complete protection or the induction of hypersensitive sites was not different in the case of this mutant fragment.
In addition to the protection and induction of hypersensitive sites in the region containing the essential ACS, there was partial protection of a second region from positions 180 to 203 or 204 at 25 ng of ORC. Moreover, there was induction of two hypersensitive sites in this region at positions 196 or 197 and 205 or 206. These hypersensitive sites became visible at 12.5 ng of ORC, whereas the sites associated with the essential ACS were visible at 5 ng. This second protected region is present in both the 9 of 11 match mutant and the triple 10 of 11 match mutant. Despite the ORC binding detected in vitro, this region cannot provide ARS function (Fig. 2G and data not shown). The deletion analysis in Fig. 2 demonstrates that sequences between positions 147 and 227 contribute to ARS function, but it is not clear whether the enhanced stability of the larger fragment is due to the presence of this weak ORC binding site or some other sequence.
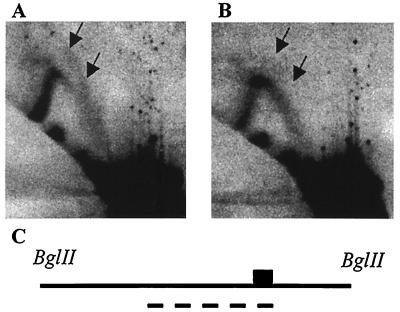
To determine whether the chromosomal replication origin associated with ARS309 depends upon the essential ACS, the chromosomal copy of ARS309 was replaced with mutant derivatives, and replication intermediates were examined by two-dimensional gel electrophoresis. Plasmids carrying mutant ARS derivatives and about 1 kb of flanking DNA were constructed and used to replace the chromosomal copy of ARS309. Fig. 4A shows the replication intermediates of a restriction fragment carrying wild-type ARS309. The presence of bubble-shaped intermediates (arrows) and the relative paucity of small Y-shaped intermediates indicate that ARS309 is active as a chromosomal replication origin. In contrast, when the Ars− mutation (Fig. 2G) was present in the chromosome, no bubble-shaped replication intermediates were detected (Fig. 4B, arrows), and there was a commensurate increase in the signal from small Y-shaped intermediates, indicating that origin function is abolished by this mutation. Strains carrying the triple mutation altering all four 10 of 11 matches (Fig. 2F) and the double mutation changing the two nonessential 9 of 11 matches (Fig. 2H) gave a pattern similar to the strain carrying wild-type ARS309 (data not shown).
Figure 4.
Analysis of ARS309 replication intermediates. Genomic DNA was digested with BglII, electrophoresed, and blotted as described by Brewer and Fangman (5). The 3.6-kb BglII fragment containing ARS309 was detected by probing with the 1.4-kb HindIII–EcoRI fragment indicated by the dashed lines in C. The 0.35-kb HpaI–EcoRI fragment containing ARS309 is denoted by the box. (A) Wild-type ARS309. The arrows point to bubble-shaped replication intermediates, indicating that ARS309 is active as an origin on the chromosome. (B) ARS309 with the essential 9 of 11 match mutated (Fig. 2G). There are no bubble-shaped replication intermediates (arrows), demonstrating that origin function is abolished in the mutant.
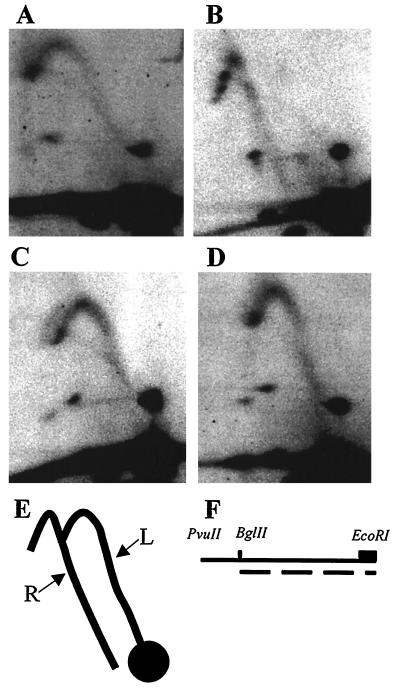
An analysis of the direction of replication fork movement (40) through the regions flanking ARS309 was performed on these mutants to determine whether there was any small reduction in origin usage that might have been missed by the standard two-dimensional gel analysis in Fig. 4. The region immediately to the left of ARS309 was examined as a 3.5-kb PvuII–EcoRI fragment that was cleaved in situ with BglII prior electrophoresis in the second dimension (Fig. 5F). When ARS309 was active (Fig. 5 A and E), replication forks moved leftward from ARS309 at the extreme right end of this fragment, resulting in a fork-direction pattern in which the early replication intermediates originate from the position of the unreplicated BglII–EcoRI fragment. In contrast, when ARS309 was inactive (Fig. 5 B and E), replication forks originating from ARS307 or ARS308 moved rightward through this region, resulting in a pattern in which the early replication intermediates arose from a position to the left of the unreplicated BglII–EcoRI fragment. The presence of rightward-moving replication forks in the Ars− mutants confirms and extends the conclusion that the 2-bp substitution that inactivates ARS309 also inactivates its activity as a chromosomal replicator.
Figure 5.
Direction of replication fork movement in the left-flanking region of ARS309 and mutant derivatives. Genomic DNA was digested with PvuII and EcoRI before electrophoresis in the first dimension. Gel slices were excised, and the DNA was digested in situ with BglII prior to electrophoresis in the second dimension. (A) Wild-type ARS309. (B) Essential ACS mutant (Fig. 2G). (C) ARS309 with all 10 of 11 matches mutated (Fig. 2F). (D) ARS309 with nonessential 9 of 11 matches mutated (Fig. 2H). (E) Diagram of replication intermediates. When ARS309 is active, leftward-moving replication forks traverse the EcoRI–PvuII fragment (see F), and replication intermediates fall on the arc labeled L. When ARS309 is inactive, rightward-moving forks from ARS307 or ARS308 traverse the fragment, and the replication intermediates fall on the arc labeled R. (F) Line drawing indicating the position of relevant restriction sites and of the 0.35-kb HpaI–EcoRI fragment containing ARS309 (box). The 2.7-kb EcoRI–BglII fragment used as a probe is represented by the dashed line.
We also examined a mutant in which all four 10 of 11 matches were eliminated (Fig. 2F) and a mutant in which the two nonessential 9 of 11 matches were eliminated (Fig. 2H); both gave patterns indistinguishable from wild-type, indicating that these mutations have no effect on origin function (Fig. 5 C and D).
The direction of replication fork movement through the region to the right of ARS309 was also examined. As we reported previously for a small circular derivative of chromosome III (36), forks traverse this region from right to left regardless of the activity of ARS309, so this region is not informative (data not shown).
This analysis of the ARS309 chromosomal replicator demonstrates that ARS309 is atypical, depending for activity upon an ACS that agrees with the consensus at only 9 of 11 positions. Moreover, the T → C change at position 9 present in the essential ACS inactivates other ARS elements. How then does ARS309 function? Three observations suggested a solution to us. (i) Our in vitro analysis of ARS309 indicates that ORC binds to the essential ACS. (ii) Linker-substitution mutagenesis of ARS307, ARS1, ARS121, H4 ARS, and ARS305 indicates that domain A, the region essential for ARS function, extends several base pairs to either side of the ACS (12–16, 41). (iii) Some mutations within the B1 element of ARS1 and ARS307 decrease ORC binding (27, 28), suggesting that the ORC recognition sequence is larger than the ACS.
In an attempt to identify additional sequences important for ARS function, we aligned sequences flanking the essential match to the ACS for all those ARS elements where the essential match had been identified by mutation (Table 2). There is substantial conservation of the 3 bp that flank both sides of the essential match to the ACS. On the 5′ side of the T-rich strand of the ACS, at position −1, all of the sequences have an adenine or thymine. There is also a strong bias for adenine or thymine at positions −2 (80% of ARS elements) and −3 (85% of ARS elements). Positions −4 to −6 show less bias, with 60–65% of ARS elements having an adenine or thymine at these positions. On the 3′ side of T-rich strand of the essential match, 75% of sequences have a guanine at position +1. Single-base changes at this position inhibit, but do not abolish, the activity of ARS307, ARS305, and HO ARS (15, 18, 20). Positions +2 and +3 both have thymines in 65% of sequences. This analysis suggests that the ACS should be expanded to the 17-bp sequence WWWWTTTAYRTTTWGTT, which we will refer to as the extended ARS consensus sequence (EACS).
Table 2.
Alignment of ARS sequences by essential ACS matches
| ARS | 5′ | Essential match | 3′ |
|---|---|---|---|
| ARS309 | TACATT | GTTTATATCTT | GTTTTGAAGTTTTAGCTTTG |
| ARS121 (42) | GTGAGA | TGTTTTGTTTA | ACATTAGTTTCAAATTAACA |
| ARS1 (43) | TACAGA | TTTTATGTTTA | GATCTTTTATGCTTGCTTTT |
| rDNA ARS (44) | TTGATT | GTTTATGTTTT | GTGTGATGATTTTACATTTT |
| HO ARS (20) | GTAAAA | TTTAATATTTT | GGATGAAAAAAACCATTTTT |
| H4 ARS (41) | ATTTAT | TTTTATGTTTT | GTATTTGGATTTTAGAAAGT |
| HMRE ARS (45) | AACATT | TTTTATATTTA | GGTATTAAATTGCGATTTAA |
| ARS305 (15) | TGGTTT | TTATATGTTTT | GTTATGTATTGTTTATTTTC |
| ARS307 (18. 46) | TTTTTT | ATTTATGTTTT | CTTCTTCACACATGGGTTAC |
| ARS601 (36) | GTACTA | ATTTCCATTTT | TGAGTTTAGGCGGTGCTTTC |
| ARS602 (36) | ACCAAT | TTATACGTTTA | GTTTTAACATCATCACAATG |
| ARS603 (36) | CCCTAA | TTTCATATTTT | GTATCAAAACTTTAAAATCT |
| ARS603 (36) | CCAGAT | TTTAAAGTTTT | GATACAAAATATGAAATTAG |
| ARS604 (36) | TGTTCA | TTTTACGTTTT | GTTACACAAATCCAATCGAA |
| ARS605 (36) | AGAATT | AATTACGTTTT | GTTGCTAAAGGAAACTTTAC |
| ARS606 (36) | ATATAT | ATTTATATTTT | CGTTGCAATTTCGCAATTTT |
| ARS607 (36) | TGTCTT | GTTTATATTTA | GTTACGTTGGGATCAAGTTT |
| ARS608 (36) | TTTTTT | TTTTACTTTTA | GTTTTCTTCTATGCGCAAGC |
| ARS609 (36) | CCATTT | TTTTATGTTTT | TTCCGGAATTGGCTAAATTC |
| ARS1413 (37) | GAAACA | ATTTGTATTTA | GTTGGTGTGACCTCAAATTT |
| Consensus | ---WWW | WTTTAYRTTTW | GTT----W-W----R-WT-- |
W, A or T; Y, T or C; R, A or G.
We searched the sequence of the 350-bp ARS309 fragment for matches to the EACS. The best match (positions 59–75, ATTgTTTATATcTTGTT) contains the essential match to the ACS identified by mutation. Thus, the 17-bp EACS correctly identifies the essential sequence for ARS309 function, whereas the conventional 11-bp ACS does not. A similar analysis was performed with the sequences of restriction fragments carrying the other ARS elements in Table 2. The sequences analyzed varied from 120 to 1400 bp, with an average size of 640 bp. The EACS does a better job of identifying the essential sequence than the conventional ACS does. For example, the 640-bp fragment carrying ARS608 has three 10 of 11 matches to the ACS (37). The best match to the EACS correctly identifies the essential match. Similarly, for the rDNA ARS, there is one perfect match to the ACS and two 10 of 11 matches; one of the 10 of 11 matches is essential for function (44). The best match to the EACS corresponds to the essential match. For ARS1, there is one perfect match to the ACS that is essential for function (19). However, there are four 15 of 17 matches to the EACS, one of which corresponds to the essential match. Even in this case, the EACS identifies the correct match as a candidate. Overall, the EACS correctly identifies the essential match to the ACS in 14 of the 20 sequences. In three other cases, the EACS identifies the correct match, but also one or more sequences with the same number of mismatches. In contrast, the conventional ACS only identifies 12 essential matches correctly, and in five other cases, the correct match is one of several with the same number of mismatches. We are using the 17-bp EACS to guide our analysis of other ARS elements from chromosomes III of S. cerevisiae and Saccharomyces carlsbergensis and have found it to be a better predictor of essential sequences than the original ACS, identifying the essential ACS in four of six ARS elements examined (C. Yang, J.F.T., and C.S.N., unpublished results).
The EACS also identifies a 14 of 17 match at positions 167–183 of the ARS309 sequence that may account for the weak ORC binding noted in Fig. 3. In addition, the weak protection of positions 159, 165, and 167 may also result from ORC binding at this EACS. The hypersensitive sites at positions 195–196 and 204–205 are spaced the same distance from this sequence as the hypersensitive sites at positions 87 and 97 are from the best match to the EACS. However, there are two other 14 of 17 matches that fail to bind ORC (positions 98–114 and 205–221).
In addition to the conserved sequences that we propose are part of an expanded ACS, the sequence comparison in Table 2 revealed five other conserved positions. The base at positions +8, +10, and +17 is adenine or thymine in 85%, 80%, and 90% of the sequences, respectively. Position +15 is a purine in 85% of the sequences, and position +18 is thymine in 75% of the sequences. These positions are indicated in Fig. 1 for the three ARS elements that have been analyzed by linker-substitution mutagenesis. Positions +17 and +18 are within the B1 elements of all three of these ARS elements (12–14, 47). In fact, mutations at position +18 of ARS1 diminish ORC binding (28). Positions +10 and +15 fall within the B1 elements of ARS1 and ARS305 but not within that of ARS307, which is further away from the ACS. Thus, the conservation of DNA sequence at these positions likely identifies base pairs of functional importance. The reason for conservation at position +8 is less clear. Position +8 lies between the ACS and the B1 elements of ARS1, ARS305, and ARS307, a region where mutations have no effect on ARS function. Further experiments will be required to determine whether this position is important in other ARS elements.
In summary, the identification of an atypical ACS in ARS309 has led to a reassessment of the sequences required for ARS function. It seems likely that the detailed analysis of additional ARS elements will reveal features that are not apparent from the small sample currently available.
Acknowledgments
We thank Lisa Macera-Bloch and Hansen Su for help with constructing mutant derivatives of ARS309, Dr. Stephen Bell for recombinant ORC and advice on footprinting, and Drs. Michael Newlon and Lynn Ripley for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM35679 to C.S.N.
ABBREVIATIONS
- ARS
autonomously replicating sequence
- ACS
ARS consensus sequence
- ORC
origin recognition complex
- EACS
extended ARS consensus sequence
References
- 1.Theis J F, Newlon C S. In: The Mycota. Brambl R, Marzluf G A, editors. Vol. 3. Berlin: Springer; 1996. pp. 1–28. [Google Scholar]
- 2.Hsiao C L, Carbon J. Proc Natl Acad Sci USA. 1979;76:3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stinchcomb D T, Struhl K, Davis R W. Nature (London) 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 4.Huberman J A, Spotila L D, Nawotka K A, el-Assouli S M, Davis L R. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 5.Brewer B J, Fangman W L. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 6.Deshpande A M, Newlon C S. Mol Cell Biol. 1992;12:4305–4313. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivier D H, Rine J. Science. 1992;256:659–663. doi: 10.1126/science.1585179. [DOI] [PubMed] [Google Scholar]
- 8.Huang R Y, Kowalski D. EMBO J. 1993;12:4521–4531. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marahrens Y, Stillman B. EMBO J. 1994;13:3395–3400. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubey D D, Davis L R, Greenfeder S A, Ong L Y, Zhu J G, Broach J R, Newlon C S, Huberman J A. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newlon C S, Collins I, Dershowitz A, Deshpande A M, Greenfeder S A, Ong L Y, Theis J F. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 12.Marahrens Y, Stillman B. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 13.Rao H, Marahrens Y, Stillman B. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theis J F, Newlon C S. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R Y, Kowalski D. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker S S, Malik A K, Eisenberg S. Nucleic Acids Res. 1991;19:6255–6262. doi: 10.1093/nar/19.22.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broach J R, Li Y Y, Feldman J, Jayaram M, Abraham J, Nasmyth K A, Hicks J B. Cold Spring Harbor Symp Quant Biol. 1983;47:1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- 18.Van Houten J V, Newlon C S. Mol Cell Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Celniker S E, Sweder K, Srienc F, Bailey J E, Campbell J L. Mol Cell Biol. 1984;4:2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearsey S. Cell. 1984;37:299–307. doi: 10.1016/0092-8674(84)90326-x. [DOI] [PubMed] [Google Scholar]
- 21.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 22.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 23.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 24.Bell S P, Kobayashi R, Stillman B. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 25.Fox C A, Loo S, Dillin A, Rine J. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 26.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowley A, Cocker J H, Harwood J, Diffley J F. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao H, Stillman B. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Houten J V. Ph.D. thesis. Newark: University of Medicine and Dentistry of New Jersey; 1990. [Google Scholar]
- 30.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newlon C S, Lipchitz L R, Collins I, Deshpande A, Devenish R J, Green R P, Klein H L, Palzkill T G, Ren R B, Synn S, Woody S T. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 33.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 34.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 35.Richterich P, Lakey N D, Lee H M, Mao J I, Smith D, Church G M. Nucleic Acids Res. 1995;23:4922–4923. doi: 10.1093/nar/23.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenfeder S A, Newlon C S. Mol Biol Cell. 1992;3:999–1013. doi: 10.1091/mbc.3.9.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirahige K, Iwasaki T, Rashid M B, Ogasawara N, Yoshikawa H. Mol Cell Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman K L, Diller J D, Ferguson B M, Nyland S V, Brewer B J, Fangman W L. Genes Dev. 1996;10:1595–1607. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- 39.Newlon C S, Theis J F. Curr Opin Genet Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- 40.Friedman K L, Brewer B J. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 41.Bouton A H, Smith M M. Mol Cell Biol. 1986;6:2354–2363. doi: 10.1128/mcb.6.7.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker S S, Francesconi S C, Eisenberg S. Proc Natl Acad Sci USA. 1990;87:4665–4669. doi: 10.1073/pnas.87.12.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Celniker S E, Campbell J L. Cell. 1982;31:201–213. doi: 10.1016/0092-8674(82)90420-2. [DOI] [PubMed] [Google Scholar]
- 44.Miller C A, Kowalski D. Mol Cell Biol. 1993;13:5360–5369. doi: 10.1128/mcb.13.9.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brand A H, Micklem G, Nasmyth K. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- 46.Palzkill T G, Newlon C S. Cell. 1988;53:441–450. doi: 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- 47.Huang T, Campbell J L. J Biol Chem. 1995;270:9607–9614. doi: 10.1074/jbc.270.16.9607. [DOI] [PubMed] [Google Scholar]