Abstract
Differential rates of nucleotide substitutions among taxa are a common observation in molecular phylogenetic studies, yet links between rates of DNA evolution and traits or behaviors of organisms have proved elusive. Likelihood ratio testing is used here for the first time to evaluate specific hypotheses that account for the induction of shifts in rates of DNA evolution. A molecular phylogenetic investigation of mutualist (lichen-forming fungi and fungi associated with liverworts) and nonmutualist fungi revealed four independent transitions to mutualism. We demonstrate a highly significant association between mutualism and increased rates of nucleotide substitutions in nuclear ribosomal DNA, and we demonstrate that a transition to mutualism preceded the rate acceleration of nuclear ribosomal DNA in these lineages. Our results suggest that the increased rate of evolution after the adoption of a mutualist lifestyle is generalized across the genome of these mutualist fungi.
Keywords: lichen symbiosis, nucleotide substitution rate, Omphalina, phylogenetic comparative method, ribosomal DNA
Mutualistic associations between fungi and photoautotrophic organisms (lichens and mycorrhizae) are almost ubiquitous in nature (1, 2). These symbiotic associations are concentrated in specific groups of fungi and are often tied to the diversification of at least one of the symbionts, suggesting that mutualism plays a major role in the evolution of most plants and in more than one-fifth of all known fungi (2–6). To study prerequisite conditions for, and evolutionary consequences of, a transition to mutualism in fungal lineages, a model system was developed (7) consisting of a group of closely related lichen-forming and nonmutualistic species of the mushroom genus Omphalina Quél. (Fig. 1a–d).
Figure 1.
Examples of mutualist (a–g) and nonmutualist fungal species (h–j) included in the present study. Mutualist lichen-forming basidiomycetes associated with the unicellular green alga Coccomyxa (a–e). (a) Omphalina hudsoniana: the green squamule-like structures covering the soil, forming the thallus of this lichen, are comprised of the fungal and algal symbionts (34). The yellow mushrooms are the sexual reproductive structures of the fungal partner of this lichen. (b–d) For these three lichen species, the thallus consists of a green crust covering the substrate (not visible in d). Their thallus is comprised of small globular structures containing the algal symbiont (34). (b) O. luteovitellina (Pilát & Nannf.) M. Lange, (c) O. velutina (Quél.) Quél., (d) O. ericetorum. (e) The thallus of this coral lichen-forming fungus, Multiclavula mucida (Fr.) R. Petersen, is not as differentiated as the one of lichen-forming Omphalina species. It forms the continuous green crust covering the substrate in this picture. (f and g) Mutualist mushroom species associated with thallose liverworts Marchantia polymorpha and Blasia pusilla, respectively: (f) Gerronema marchantiae, (g) Rickenella pseudogrisella. (h–j) Closely related nonmutualist mushroom species: (h) R. mellea (Sing. & Clémenç.) Lam., (i) O. velutipes Orton, (j) Arrhenia auriscalpium (Fr.) Fr. Photographs are by Kolbjørn Mohn Jenssen (a–d and f–j) and Irwin Brodo (e); the former are reproduced with the permission of G. Gulden and K. M. Jenssen (35–37).
Faster rates of nucleotide substitutions have been observed in endosymbiotic bacteria associated with aphids compared with free-living bacteria (8, 9). Mitochondria in fungi and animals also are reported to evolve faster than related bacteria (10). From these observations, it is tempting to conclude that accelerated rates of nucleotide substitutions resulted from an evolutionary transition to a symbiotic state. This conclusion is dependent on the sampling of free-living and symbiotic taxa, as well as on the extent of nucleotide sequence sampling. Both factors have a direct effect on the degree of confidence associated with a given phylogenetic tree and estimates of rates of nucleotide substitutions. Moreover, because nucleotide sequences contain phylogenetic structure, pairwise sequence comparisons are not independent, and thus a simple χ2 test of association may overestimate the true strength of any association between evolutionary rates and symbiosis (11–13). Tests that take account of phylogenetic relationships are required for a better understanding of the factors causing shifts in rates of DNA evolution. Here we investigate, using a phylogenetically based maximum likelihood procedure, the association between transitions to mutualism and shifts in rates of nucleotide substitutions, as well as their relative order of occurrence, in a group of free-living and mutualist fungi (ref. 13 and M.P., unpublished work).
MATERIALS AND METHODS
Phylogenetic Analyses.
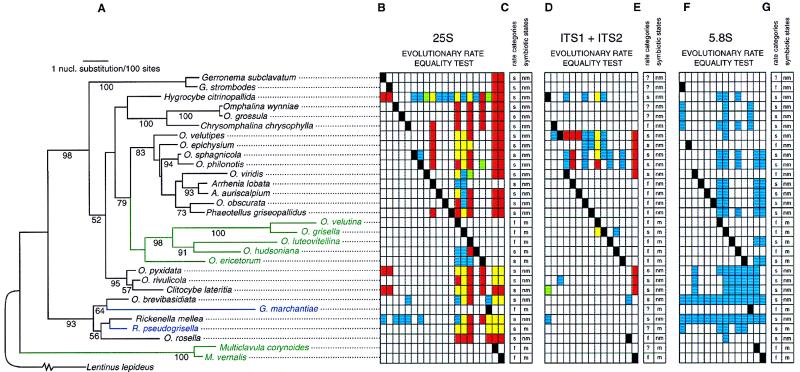
The phylogenetic tree of Fig. 2 was estimated using fastdnaml Ver. 1.1.1a (14, 15) on a combined data set including a total of 1550 sites from the nuclear ribosomal DNA (nrDNA) 25S, 5.8S, ITS1, and ITS2 (16). One hundred independent local rearrangement searches were performed followed by global rearrangement searches on all different topologies generated by the 100 local rearrangement searches. The fastdnaml searches on the combined data set were performed with four different categories of evolutionary rates for each of the four molecular data sets (5.8S = 0.60, 25S = 0.92, ITS1 = 1.68, and ITS2 = 1.96). Gap poor regions 1–6 of ITS1 and 1–5 of ITS2 (16) constituted the ITS1+ITS2 data set used in the present study. Readers should refer to Lutzoni (16) for detailed descriptions of methods used for the alignment, tests for phylogenetic signal, tests for combinability of multiple data sets, and phylogenetic analyses that lead to the phylogenetic tree shown in Fig. 2.
Figure 2.
(A) Phylogenetic relationships (ln likelihood = −7370.71124) of 30 species of Omphalina and related genera. The number below each internode is the percentage of 350 bootstrap replicates supporting the same binary partition as shown in this figure. Boostrap values ≤50% are not shown. In green are lichen-forming fungal lineages associated with the green alga Coccomyxa. In blue are fungal lineages mutualistically associated with thallose liverworts. (B, D, and F) Each column represents all pairwise comparisons between one species (species “a,” black box) and the remaining 29 species (species “x,” nonblack boxes). A white box indicates that the evolutionary rate of species x does not differ from the rate of species a. A colored box indicates that species x is significantly (P < 0.05) slower than species a. Significant differences in rates of nucleotide substitution were subdivided into four types: (i) Blue boxes represent cases in which rate differences are due to a significant difference in rates of transitions only; (ii) yellow boxes indicate that rate differences are due to significant differences in rates of transitions and rates of transversions; (iii) red boxes represent cases in which the rate differences are due to significant differences in rates of transversions only; and (iv) green boxes represent cases in which the rate difference is not due to a significant difference in rates of transitions nor rates of transversions when tested separately. For example, in the first column (B), the evolutionary rate of Gerronema subclavatum (Peck) Redhead was not significantly faster than G. strombodes (Berk. & Mont.) Sing. The evolutionary rate of G. subclavatum was significantly faster than Hygrocybe citrinopallida (Smith & Hesler) Kobay, and this significant rate difference is due to a significant difference in rates of transversions only. The presence of red and yellow boxes for the 25S and ITS1+ITS2 (B and D) emphasizes their fast evolutionary rates, compared with the slowly evolving 5.8S, which is entirely blue (F). For space considerations, columns were not created for species that were not significantly faster than any other taxon, i.e., taxa with no black box in their respective row (e.g., Hygrocybe citrinopallida). (C, E, and G) Evolutionary rate categories (f = fast, s = slow) and symbiotic states (m = obligatory mutualist, nm = nonmutualist) represent the two characters tested for correlated evolution by one of the two analyses used in this study (see Table 1 for the results of this test).
The support for the internodes of the maximum likelihood tree was estimated by 350 bootstrap replicates. Bootstrapped data sets were generated using the seqboot program in phylip (17). One global search was done on each bootstrapped data set using fastdnaml with four rate categories (as described above), the jumble option, and a transition/transversion ratio of 2.14. The 350 searches were performed using the “P4” code, which distributed the computational task to 63 processors in parallel on an Intel Paragon supercomputer. The bootstrap values were computed for the resulting 350 bootstrapped trees using the majority rule consensus option of paup 3.1.1.
The gains and losses of mutualism, as well as the evolution of each 1550 nrDNA nucleotide positions, were reconstructed using macclade Ver. 3.05 (18). Unambiguous nucleotide substitutions in the mutualist clade and nonmutualist sister group were recorded as disrupting, creating, or neutral to the potential formation of thymine dimers. The frequency of unambiguous nucleotide substitutions was recorded for each adjacent nucleotide pair (overlapping) along the large subunit and 5.8S nrDNA sequences.
Testing for Equality of Evolutionary Rates.
Equality of evolutionary rates, in general and restricted to transitions or transversions, was tested using the program codrates (19). This likelihood ratio test was applied to all possible pairwise comparisons of the 30 taxa forming the ingroup, representing 435 tests. These tests were implemented on the 25S, 5.8S, and ITS nrDNA data sets separately, for a total of 1305 tests. The results of these tests are summarized in Fig. 2 B, D, and F. These tests have varying degrees of nonindependence because of shared phylogenetic branches among species. We therefore treat them as descriptive only, i.e., to determine which of the observed rate differences between two given species are statistically significant.
Phylogenetic Comparative Tests.
Shifts in evolutionary rates of the nrDNA sequences and in symbiotic states for these 30 mushroom species were investigated using two different likelihood ratio tests designed specifically to analyze the relationship between two binary characters (13) or between one continuous and one binary character (M.P., unpublished work) that are measured across a group of hierarchically evolved species or populations. The first method (13) allows one to analyze the evolutionary association between the two binary characters (i.e., do they tend to co-evolve?) and to test whether the changes in one character precede those in the other, all while taking account of the phylogenetic relationships and making use of branch length information in the phylogeny. We used this method to test for an association between evolutionary transitions to mutualism and shifts from slow to fast evolutionary rates and to ask whether evolutionary transitions to mutualism preceded or followed transitions to faster rates of evolution. The method required the characterization of each species as fast or slow evolving based on the 25S, 5.8S, and ITS nrDNA data sets separately. Species were categorized as fast “f” when they were found to be significantly faster than at least one species and not significantly slower than any other species (represented by a row with a black box and no colored box in Fig. 2 B, D, or F). The remaining species were classified as slow “s.” The association between the changes observed in the two binary characters (rate categories and symbiotic states; Fig. 2 C, E, and G) over the entire phylogenetic tree (Fig. 2A) was then estimated using the continuous time Markov model with the program discrete (13).
The second method (M.P., unpublished work) allows a direct test of whether mutualists are associated with increased evolutionary rates without relying on categorizing species into slow or fast evolvers. The association between branch length estimates derived from maximum likelihood (15) and symbiotic state was assessed by a likelihood ratio statistic (M.P., unpublished work). Using branch length estimates (in units of expected number of nucleotide substitutions per site) from the maximum likelihood tree, the statistic estimates a single rate of evolution parameter for the entire tree and then compares the fit of that model to one in which two different rate parameters are fit, one for each symbiotic state. If the branch lengths corresponding to the two symbiotic states differ from one another, the two-parameter model will give a better fit to the data. The key feature of the model is how it deals with the problem of what the ancestral character states are for the symbiotic state character. The method fits the two-rate parameter models while simultaneously varying the ancestral states over all possible values [using the same logic as described by Pagel (13)]. This means that the resulting rate parameters do not depend on any one reconstruction of the ancestral character states.
RESULTS
Evolutionary Transitions to Mutualism.
A molecular phylogenetic study of 17 species of Omphalina and 13 species selected from putative related genera, based on nrDNA sequences, revealed at least four independent transitions to mutualism (16). One of these evolutionary transitions gave rise to a monophyletic group of five lichen-forming Omphalina species (Fig. 2A). A second transition gave rise to the lichen-forming genus Multiclavula R. Petersen, represented by two species in this study (Figs. 1e and 2A). Both lichen-forming Omphalina and Multiclavula species are mutualistically associated with the unicellular green alga Coccomyxa W. Schmidle. The mutualistic fungal species Gerronema marchantiae Sing. and Clémenç. and Rickenella pseudogrisella (A. H. Smith) Gulden, which are associated with thallose liverworts Marchantia L. and Blasia pusilla L., respectively, represent the two other independent transitions to mutualism (Figs. 1 f and g and 2A).
Maximum Likelihood Ratio Test of Equality on Evolutionary Rates from the 25S Data Set.
Branch lengths of the phylogenetic tree for these 30 ingroup species (Fig. 2A) suggested that an increase in the rate of nucleotide substitution was associated with a transition to mutualism. To determine which pairwise differences in rates of nucleotide substitution were statistically significant, a maximum likelihood ratio test of equality on evolutionary rates (19) was performed on all possible pairs of species. Of the 435 possible pairwise comparisons between species, 145 significant differences (P < 0.05) in rates of nucleotide substitutions were detected based on 1264 nucleotide positions from the 25S nrDNA (Fig. 2B). Of these, 107 (74%) were mutualist fungal (m) species evolving at faster rates (f) than nonmutualist (nm) species, 26 (18%) were comparisons between two nonmutualistic fungal species, and 8% involved comparisons between two mutualistic fungal species. These pairwise tests among species are not independent because they share branches of the phylogenetic tree and because, even if they were phylogenetically independent, not all of the possible pairwise tests can be mutually exclusive.
Phylogenetic Comparative Tests of the Large Subunit, ITS, and 5.8S nrRNA.
There was an almost perfect correspondence between mutualism (m) and a fast rate of nucleotide substitution (f), except for Rickenella pseudogrisella, Omphalina hudsoniana (Jennings) Bigel., and O. ericetorum (Fr.) M. Lange, which were categorized as slowly (s) evolving mutualists. However, the two latter mutualist species were only found to be slower than other faster mutualist Omphalina species (Fig. 2 B and C). The slow evolutionary rate of R. pseudogrisella could be explained by a very recent transition to mutualism and, therefore, is not in contradiction with an accelerated evolutionary rate induced by a transition to mutualism. All species categorized as evolving at a fast rate (f) were mutualists (Fig. 2 B and C).
Two phylogenetic comparative tests were applied, and both revealed that mutualism and evolutionary rate are correlated based on 25S data (see Table 1: likelihood ratio = 18.6, P < 0.001; and Table 2: likelihood ratio = 15.9, P = 0.02). The high rate of change from a mutualist with a slow rate of evolution (m,s) to a mutualist with a fast rate of evolution (m,f) (q34 = 1.180826) compared with the low rate of change from a nonmutualist with a slow rate of evolution (nm,s) to a nonmutualist with a fast rate of evolution (nm,f) (q12 = 0.001074) obtained from the correlated-change analysis (Table 1) shows that transitions to a fast rate of nucleotide substitution are more likely to occur in mutualistic lineages than in nonmutualistic lineages (Table 1: q34 > q12, P < 0.00001). Furthermore, the low rate of change from nm, s to nm, f (q12 = 0.001074) compared with the higher rate of change from nm,s to m,s (q13 = 0.091419) shows that transitions to mutualism precede transitions to a fast rate of nucleotide substitutions in the evolutionary history of these lineages. This trend was found also to be highly significant (Table 1: q13 > q12, P < 0.00001). Thus, we find a strong association between rates of evolution and symbiotic state, such that higher rates of nucleotide substitutions in the 25S occur after transitions to mutualism.
We tested for an association between evolutionary rates and symbiotic state categories in the two ITS regions and the 5.8S nrDNA (Fig. 2D–G) using the same likelihood methodologies (13, M.P., unpublished work) as for the 25S data. With both methods, the association was found to be significant for the ITS regions (P = 0.05, Table 1; P = 0.03, Table 2) and nearly so for the 5.8S molecule (P = 0.07, Table 1; P = 0.07, Table 2).
DISCUSSION
The increased rates of nucleotide substitutions seen among mutualistic Omphalina species could result from relaxation of selection, positive selection on coding regions of the nrDNA, or a generalized genomic acceleration of nucleotide substitutions. Analysis of four regions of the nrDNA repeat unit points to the latter as the most probable cause.
Relaxation of selection is commonly associated with increased A + T concentrations (20–24). The ITS1 and ITS2 regions of the nrDNA are removed by splicing during intermediary stages of rRNA maturation and do not bind with proteins to form ribosomes. It is therefore assumed that these regions are under less selective pressure than the 25S and 5.8S genes, which are integral parts of the ribosomal large subunit. For the 30 ingroup species included in this study, the two ITS regions did evolve at faster rates (ITS1 = 1.7 changes/site, ITS2 = 2.0 changes/site) relative to the 25S (1.0 changes/site) and 5.8S (0.6 changes/site), and they had a slightly higher A + T content (ITS2 = 56%, ITS1 = 59% vs. 25S = 51%, 5.8S = 55%). Therefore, if the increase in rates of nucleotide substitutions in the 25S of mutualists resulted from relaxed selection, we would expect to observe an increase in A + T content in this molecule of the mutualists. However, we did not observe such a pattern in the 25S when comparing the base composition between nonmutualists and mutualists (A + T = 51% for nonmutualists vs. 50% for mutualists). Instead, the rate increase in the lichen-forming Omphalina clade compared with its nonmutualistic sister group was due to an increase in A-to-G substitutions (16.4% vs. 7.7%), T-to-C (26.6% vs. 21.9%), T-to-A (6.3% vs. 1.3%), A-to-T (5.5% vs. 1.3%), and T-to-G (3.1% vs. 1.3%).
To distinguish between increased levels of selection on coding regions and a generalized genomic increase in nucleotide substitutions to account for the increase in mutualists’ evolutionary rates, we examined the pattern of rate increase in conserved regions of the ITS1 and ITS2 (151 nt positions) and the entire 5.8S gene (161 nt positions) of the nrDNA for the same 30 species (Fig. 2 D and F). If faster substitution rates of mutualists were generalized for the genome and were not caused by differential selection, the association between fast evolutionary rates and mutualism observed in the 25S molecule (Tables 1, 2) should also be observed in these other sequences. As for the 25S, both comparative tests detected a significant association between transitions to mutualism and accelerated rates of evolution for the ITS data set and nearly so for the 5.8S gene. The latter is a short and slowly evolving segment of the nrDNA, so slightly weaker associations are expected for it. These results, therefore, do not support the hypothesis that positive selection on the coding regions of the nrDNA is the cause for the observed shift in evolutionary rates associated with a transition to mutualism but rather suggest a generalized increase in rates of nucleotide substitutions in these symbiotic species.
A generalized genomic acceleration of nucleotide substitutions in mutualistic species could reflect: a genetic bottleneck that took place in a highly genotypically variable population, leading to mutualist groups; deficient DNA repair systems (25); shorter generation time (26); reduced sexual reproduction (3) coupled with smaller population size (27); and an increase of mutagenic free radicals in the mycobiont caused by increased levels of oxygen, dessication, ionizing radiation, or UV-A and UV-B radiations between 320 and 380 nm (28–30) or UV radiation as a direct generalized selective force causing the formation of cyclobutane pyrimidine dimers (31, 32). The results of this study point to the two latter hypotheses as the best explanations.
The signature of a severe genetic bottleneck taking place in a highly genotypically variable population causing a burst of fixation that would be associated with a transition to mutualism would be a long internode supporting the lichen-forming Omphalina clade; however, the short length of this internode does not support this hypothesis (Fig. 2A). It is possible that one of the DNA repair systems might be deficient in lichen-forming Omphalina because meiotic anomalies were observed (by light microscopy) in these species (7, 33). However, at this time, there are no data available to explore the hypothesis that the DNA repair systems of these mutualist species are somehow deficient nor is there any evidence suggesting that a reduction in generation time is associated with these mutualist species. The loss of sexual reproduction has been proposed as a consequence of transitions to mutualism (3), and Muller’s rachet within small asexual populations has been reported as the best explanation for accelerated evolution of endosymbiotic bacteria (27). Lichen-forming species of Omphalina and Multiclavula do reproduce asexually with vegetative propagules that contain both symbionts. It is possible that asexual reproduction for these mutualist fungal species contributes much more (if not entirely) to their overall reproduction than sexual reproduction (7, 33). It is also possible that all speciation events after a transition to mutualism in the genus Omphalina were associated with genetic bottlenecks in one of the two diverging lineages, which would explain this fractal asymmetric pattern of faster lineages as sister group to slower lineages throughout the lichenized Omphalina clade (Fig. 2A). However, more evidence is needed to confirm the weak efficiency of sexual reproduction in the lichen-forming lineages; and this hypothesis does not account for the fast rate of evolution of Gerronema marchantiae, which is not known to reproduce asexually.
The two following hypotheses best explain the accelerated rates of evolution that were detected subsequent to transitions to mutualism in these fungal lineages. The transfer of the mycelium from beneath the substrate surface (Fig. 1 h-j) to growing on the substrate surface with an alga or liverwort (Fig. 1 a-g), as a direct consequence of a transition to mutualism, exposed the mycelium to desiccation and solar radiation. Furthermore, the fungal hyphae were growing in direct contact with photosynthetic cells. Both desiccation and higher levels of ambient oxygen could induce the production of superoxides and other mutagenic free radicals (28–30) in the fungal symbiont, resulting in increased mutation rates.
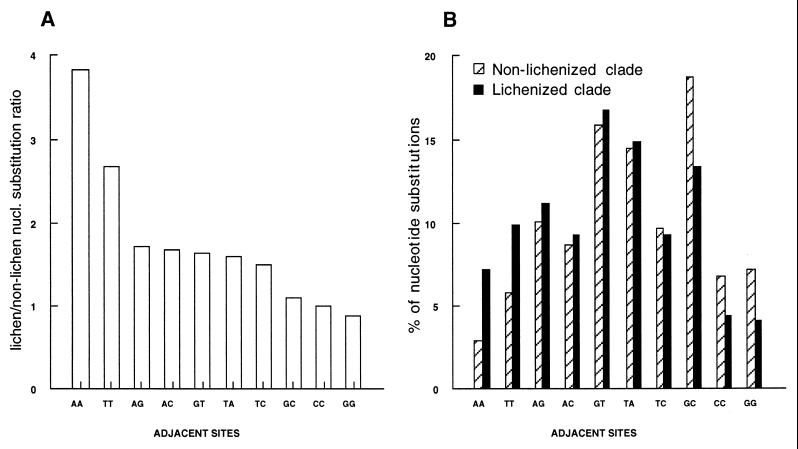
A second, nonexclusive scenario that could account for a general increase in rates of nucleotide substitutions invokes UV radiation as the selective force (31, 32). Pyrimidine dimers are known to form after the interaction of UV radiation with DNA. These dimers can inhibit future DNA replication (30). One of the major consequences of a transition to a lichen-forming state in these fungal lineages is the exposure of the mycelium to UV radiation. A comparison of all nucleotide substitutions in the lichen-forming Omphalina clade vs. the nonmutualist sister group indicated that 31% (vs. 20%) of the changes eliminated the potential formation of thymine dimers. Only 14% (vs. 22% in the nonmutualist sister group) created the potential formation of new thymine dimers whereas all of the other changes (55% vs. 58%) were neutral in this regard. The accelerated rate of nucleotide substitution of the lichenized Omphalina clade, when compared with its sister group, was caused mainly by a specific increase of nucleotide substitutions at AA and TT sites in the lichenized lineages (Fig. 3). Of interest, most of the nucleotide substitutions were occurring at GT, TA, and GC sites for both clades. This trend associated with potential sites of thymine dimerization was not observed when comparing the mutualist Gerronema marchantiae to the nonmutualist sister species Omphalina brevibasidiata (Sing.) Sing. and, therefore, cannot account for the accelerated evolution of G. marchantiae.
Figure 3.
Frequency of nucleotide substitutions along adjacent nucleotide sites for the large subunit and 5.8S nrRNA genes. The comparison is between the lichenized Omphalina clade (five species) and its sister nonlichenized clade (nine species; Fig. 2A). (A) Number of nucleotide substitutions in the lichenized clade/number of nucleotide substitutions in the nonlichenized sister clade for each adjacent nucleotide pairs. (B) Distribution of nucleotide substitutions (%) for all types of adjacent nucleotide sites as recorded for the lichenized and nonlichenized sister clades.
These results strongly suggest that two different driving forces account for the generalized acceleration in rates of evolution in the mutualist species included in this study. The generalized genomic acceleration in the lichen-forming Omphalina lineage is mostly due to selection of mutations that disrupt the potential formation of thymine dimers. For Gerronema marchantiae, the cause of the acceleration is not known but could be caused by a generalized acceleration of mutation rates caused by the formation of mutagenic free radicals in the fungal symbiont.
Table 1.
Testing for correlated evolution between rates of nucleotide substitution and symbiotic states as binary characters
| Data set (nrDNA) | Independent analysis
|
Dependent analysis
|
Likelihood ratio | Simulation, P | q34 > q12 (P) | q13 > q12 (P) | q13 < q12 (P) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Likelihood L(I) | Likelihood L(D) | q12 nm,s → nm,f | q13 nm,s → m,s | q21 nm,f → nm,s | q24 nm,f → m,f | q31 m,s → nm,s | q34 m,s → m,f | q42 m,f → nm,f | q43 m,f → m,s | ||||||
| 25S | −33.34 | −24.02 | 0.001074 | 0.091419 | 1.813253 | 2.161151 | 0.682283 | 1.180826 | 0.000065 | 0.568961 | 18.6 | <0.001 | <0.00001 | <0.00001 | |
| ITS1+ITS2 | −33.41 | −29.50 | 0.136537 | 0.130906 | 0.310519 | 0.000194 | 1.023416 | 1.440970 | 0.000021 | 0.346760 | 7.8 | 0.05 | 0.05 | not significant | |
| 5.8S | −31.61 | −27.84 | 0.467298 | 0.079326 | 2.651783 | 0.130013 | 0.513440 | 0.782133 | 0.004209 | 0.391890 | 7.5 | 0.07 | not significant | <0.01 | |
qij represent all possible transitions of one binary character holding the state of the other constant. L(I) = likelihood value for the model of independent evolution (13), and L(D) = likelihood value for the model of correlated evolution. nm, nonmutualist; m, mutualist; s, slow rate of nucleotide substitution; f, fast rate of nucleotide substitution. To test the hypothesis that a shift in evolutionary rate is more likely to occur in a mutualist lineage, the rates of change from nm,s to nm,f (q12) and from m,s to m,f (q34) were constrained to be equal, and the likelihood of this model was compared to a model in which the rates were not constrained. To test whether a transitional change in a branch to mutualism occured first followed by an increase in evolutionary rate, a likelihood ratio test was implemented by constraining the rate of change from nm,s to nm,f (q12) and the rate of change from nm,s to m,s (q13) to be equal.
Table 2.
Testing for correlated evolution between rates of nucleotide substitutions, unconverted to a binary state character, and symbiotic states as a binary character
| Data set (nrDNA) | Rate | Rate 1 | Rate 2 | Likelihood ratio | P |
|---|---|---|---|---|---|
| 25S | 0.73 | 1.27 | 0.12 | 15.9 | 0.02 |
| ITS1+ITS2 | 0.18 | 0.35 | 0.03 | 18.7 | 0.03 |
| 5.8S | 1.21 | 2.11 | 0.40 | 13.9 | 0.07 |
Higher Rate 1 coefficients reveal a faster rate of evolution in mutualistic lineages.
Acknowledgments
We thank M. Uyenoyama and D. Armaleo for insightful discussions that led to this paper. We are very grateful to S. Muse, who provided help with testing equality of evolutionary rates. Thanks to W. L. Culberson, C. F. Delwiche, J. Felsenstein, E. Gerecke, N. Gillham, C. Laurie, B. Mishler, N. A. Moran, M. Nedball, N. R. Pace, J. D. Palmer, K. M. Pryer, P. Pukkila, M. Uyenoyama, and R. Vilgalys for their comments on various versions of this paper. We are grateful to Josée Bélisle for graphical assistance with Fig. 2. This work was supported by a National Science Foundation Dissertation Improvement Grant (DEB 93-21828), Formation de Chercheurs et l’Aide à la Recherche (Québec), the A. W. Mellon Foundation, the Mycological Society of America, the New England Botanical Club, and a Sigma Xi Grant-in-Aid to F.L. as well as by the Medical Research Council, the Nuffield Foundation, the Natural Environment Research Council, and the Biotechnology and Biological Sciences Research Council, all of the United Kingdom (M.P.). This paper is part of a doctoral dissertation completed at Duke University by F.L. under the direction of Rytas Vilgalys.
ABBREVIATION
- nrDNA
nuclear ribosomal DNA
References
- 1.Cooke R. The Biology of Symbiotic Fungi. New York: Wiley; 1977. [Google Scholar]
- 2.Hawksworth D L. Bot J Linn Soc. 1988;96:3–20. [Google Scholar]
- 3.Law R, Lewis D H. Biol J Linn Soc. 1983;20:249–276. [Google Scholar]
- 4.Pirozynski K A, Malloch D W. BioSystems. 1975;6:153–164. doi: 10.1016/0303-2647(75)90023-4. [DOI] [PubMed] [Google Scholar]
- 5.Malloch D W, Pirozynski K A, Raven P H. Proc Natl Acad Sci USA. 1980;77:2113–2118. doi: 10.1073/pnas.77.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon L, Bousquet J, Lévesque R C, Lalonde M. Nature (London) 1993;363:67–69. [Google Scholar]
- 7.Lutzoni F, Vilgalys R. Cryptogam Bot. 1995;5:71–81. [Google Scholar]
- 8.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc Lond B. 1993;253:167–171. [Google Scholar]
- 9.Moran N A, von Dohlen C D, Baumann P. J Mol Evol. 1995;41:727–731. doi: 10.1007/BF00170675. [DOI] [PubMed] [Google Scholar]
- 10.Yang D, Oyaizu Y, Oyaizu H, Olsen G J, Woese C R. Proc Natl Acad Sci USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Am Nat. 1985;125:1–15. [Google Scholar]
- 12.Harvey P H, Pagel M D. The Comparative Method in Evolutionary Biology. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 13.Pagel M. Proc R Soc Lond B. 1994;255:37–45. [Google Scholar]
- 14.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 15.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 16.Lutzoni F. Syst Bio. 1997;46:373–406. doi: 10.1093/sysbio/46.3.373. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. phylip: PHYLogeny Inference Package. Seattle: Univ. of Washington; 1993. , Ver. 3.53c. [Google Scholar]
- 18.Maddison W P, Maddison D R. macclade: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 1992. , Ver. 3.05. [DOI] [PubMed] [Google Scholar]
- 19.Muse S V, Weir B S. Genetics. 1992;132:269–276. doi: 10.1093/genetics/132.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gojobori T, Li W-H, Graur D. J Mol Evol. 1982;18:360–369. doi: 10.1007/BF01733904. [DOI] [PubMed] [Google Scholar]
- 21.Li W-H, Wu C-I, Luo C-C. J Mol Evol. 1984;21:58–71. doi: 10.1007/BF02100628. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe K H, Morden C W, Ems S C, Palmer J D. J Mol Evol. 1992;35:304–317. doi: 10.1007/BF00161168. [DOI] [PubMed] [Google Scholar]
- 23.Aoyama K, Haase A M, Reeves P R. Mol Biol Evol. 1994;11:829–838. doi: 10.1093/oxfordjournals.molbev.a040166. [DOI] [PubMed] [Google Scholar]
- 24.Hu G, Thilly W G. Gene. 1994;147:197–204. doi: 10.1016/0378-1119(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 25.Britten R J. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- 26.Kohne D E. Q Rev Biophys. 1970;33:327–375. doi: 10.1017/s0033583500004765. [DOI] [PubMed] [Google Scholar]
- 27.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halliwell B, Gutteridge J M C. Free Radicals in Biology and Medicine. Oxford: Clarendon; 1989. [Google Scholar]
- 29.Mattimore V, Battista J R. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, D.C.: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 31.Li W-H, Graur D. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer; 1991. [Google Scholar]
- 32.Singer C E, Ames B N. Science. 1970;170:822–826. doi: 10.1126/science.170.3960.822. [DOI] [PubMed] [Google Scholar]
- 33.Lamoure D. Bibl Mycol. 1993;150:155–160. [Google Scholar]
- 34.Oberwinkler F. Beih Nova Hedwigia. 1984;79:739–774. [Google Scholar]
- 35.Gulden G, Jenssen K M, Stordal J. Arctic and Alpine Fungi I. Oslo: Soppkonsulenten; 1985. [Google Scholar]
- 36.Gulden G, Jenssen K M. Arctic and Alpine Fungi II. Oslo: Soppkonsulenten; 1988. [Google Scholar]
- 37.Senn-Irlet B, Jenssen K M, Gulden G. Arctic and Alpine Fungi III. Oslo: Soppkonsulenten; 1990. [Google Scholar]