Abstract
The P element, originally described in Drosophila melanogaster, is one of the best-studied eukaryotic transposable elements. In an attempt to understand the evolutionary dynamics of the P element family, an extensive phylogenetic analysis of 239 partial P element sequences has been completed. These sequences were obtained from 40 species in the Drosophila subgenus Sophophora. The phylogeny of the P element family is examined in the context of a phylogeny of the species in which these elements are found. An interesting feature of many of the species examined is the coexistence in the same genome of P sequences belonging to two or more divergent subfamilies. In general, P elements in Drosophila have been transmitted vertically from generation to generation over evolutionary time. However, four unequivocal cases of horizontal transfer, in which the element was transferred between species, have been identified. In addition, the P element phylogeny is best explained in numerous instances by horizontal transfer at various times in the past. These observations suggest that, as with some other transposable elements, horizontal transfer may play an important role in the maintenance of P elements in natural populations.
Keywords: horizontal transfer, mobile DNA, Sophophora
Transposable elements are universal features of eukaryotic genomes and can be broadly divided into two different classes (1). Class I elements are characterized by DNA sequences with homology to reverse transcriptase and are often referred to as retrovirus-like elements or retroelements. Their mobility is achieved through an RNA intermediate. Transposition of Class II elements, such as the Drosophila elements mariner and P, is catalyzed by a transposase and occurs directly from DNA to DNA, without an RNA intermediate. The P element was first described in D. melanogaster where its mobility in the germ-line of hybrid flies is responsible for a type of hybrid dysgenesis (2). The complete, or canonical, P element is 2,907 base pairs long and has four ORFs (ORF 0–3) that encode an 87-kDa DNA-binding transposase (3). Also required for transposition are the element termini, which include flanking 31-bp perfect inverted repeats, 11-bp subterminal repeats, and unique terminal sequences comprising approximately 150 bp (4). The genomic copy number of D. melanogaster P elements varies from 0 to about 60 per genome (3). A minority of these are autonomous (transposase-competent) elements; most are internally deleted, nonautonomous (transposase-incompetent) elements. Defective P elements are generally smaller, variable in size, and derived from complete elements by internal deletions. The induction of these deletions is associated with active transposition of P elements.
When active, transposable elements can behave as natural or spontaneous mutagens, inducing a wide variety of mutations, chromosomal aberrations, or other genetic changes. Although positive effects are not precluded, both theoretical and empirical studies of transposable elements suggest that this property of transposition has a negative effect on host fitness (5–7). Nonetheless, transposable elements are maintained and can indeed spread quite rapidly in natural and laboratory populations (8, 9). Thus, the evolutionary dynamics of transposable elements is complex, reflecting a balance between increase in copy number through replicative transposition and decrease by stochastic forces and through the action of selection. The situation is further complicated by the apparent propensity of transposable elements for horizontal transfer between species (10, 11). Indeed, horizontal transfer may be essential to the long-term survival of transposable elements (12).
An advantage of using the P element as a model system for understanding transposable element evolution is that good phylogenetic hypotheses exist for many species in the genus Drosophila. The subgenus Sophophora, which includes D. melanogaster and its relatives, has been the subject of particular attention. A growing number of molecular studies provide independent tests for phylogenies based on comparative morphology, biogeography, and behavior. This paper summarizes the results of a phylogenetic survey that spans the diversity of the subgenus Sophophora, which originated between 40 million and 60 million years ago. This includes 239 partial sequences isolated from 40 species distributed among the four principal species groups of Sophophora. Detailed phylogenetic analyses for individual species groups are presented elsewhere (ref. 13; J.B.C., P. Kim, and M.G.K., unpublished work; J. García-Plannells, N. Paricio, R. de Frutos, J.B.C., and M.G.K., unpublished work) and are integrated here to provide insights into P element evolution. Among the more unexpected results is the presence of different subfamilies of the P element, differing by as much as 40% at the nucleotide level in the same genome. Whereas vertical transmission is the dominant mode of P element evolution, isolated examples of horizontal transfer between species have been identified. Examination of the P element phylogeny in light of the species phylogeny suggests that additional horizontal transfers may have occurred at various times in the past and may explain the overall structure of the P element phylogeny in Sophophora.
MATERIALS AND METHODS
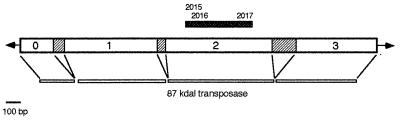
Most of the species examined in this survey were obtained from the National Drosophila Species Resource Center in Bowling Green, OH. Details concerning their actual identities and the experimental methods used are found elsewhere (ref. 13; J.B.C., P. Kim, and M.G.K., unpublished work; J. García-Plannells, N. Paricio, R. de Frutos, J.B.C., and M.G.K., unpublished work). For each species, genomic DNA was used as a template in PCR amplifications with two sets of degenerate primers specific for a region of exon 2 of the canonical D. melanogaster P element (the first P element sequenced and the standard frame of reference for subsequent sequences) (see Fig. 2). Two alternative 5′-primers are 2015, complementary to positions 1,230–1,251 of the canonical P element from D. melanogaster (3) and 2016, complementary to positions 1,305–1,327. A single 3′-primer (2017) was used complementary to positions 1,758–1,780. Primers 2015 and 2017 amplify a 550-bp fragment, and primers 2016 and 2017 amplify a 450-bp fragment. Also included in the analysis were 11 P element sequences obtained from the literature: D. subobscura A1 and G2, (14), D. guanche G1 (15), D. nebulosa N10 (16), D. willistoni L13 (17), D. bifasciata M and O types (18, 19), D. melanogaster (3), Scaptomyza pallida 2 and 18 (20), and Lucilia cuprina P1 (21).
Figure 2.
Schematic representation of the complete P element. The canonical P element from D. melanogaster is 2,907 bp in length and is flanked by perfect 31-bp inverted repeats (arrowheads). Exons 0–3 (empty boxes) encode the transposase necessary for P element mobility. The relative locations of the primers (2015, 2016, and 2017) used to amplify the DNA fragment used for phylogenetic analysis are indicated above exon 2.
The entire data set consists of the 239 partial PCR sequences and the 11 sequences mentioned above. To aid the presentation of the results, phylogenetic analysis was performed on a reduced data set comprising consensus sequences. Using macclade (22), an ancestral sequence was derived from monophyletic sequences obtained from the same species. Where ancestral nucleotides could not be assigned unambiguously, a consensus nucleotide was assigned based on the most common nucleotide present at that position in the monophyletic sequences isolated from a particular species. Species that possess more than one P element subfamily are represented by more than one consensus sequence. This reduced the total number of sequences from 250 to 80. Phylogenetic analysis was performed using parsimony as implemented by paup 3.1.1. (23). The analysis was confined to 449 characters flanked by primers 2016 and 2017, 340 of which were parsimony informative. Ten separate heuristic searches were performed on the aligned data set, each terminating after the accumulation of 1,000 equally parsimonious trees. For each search, 10 random sequence additions and tree bisection-reconnection branch-swapping were used. All 10 searches found trees of 2,089 steps, indicating that this is indeed the most parsimonious reconstruction. The data also were analyzed with the neighbor-joining algorithm (24). Using both parsimony and neighbor-joining, bootstrap analysis of 100 replicates was performed on the consensus data set.
RESULTS
Phylogeny of the Subgenus Sophophora.
To provide a frame of reference for analyzing P element evolution, the relationships among the four principal species groups of Sophophora are shown in Fig. 1, along with time estimates for major divergence events. These estimates are based on several morphological, cytological, biogeographical, behavioral, and molecular studies (see ref. 25). Although some of the divergence events and times depicted are based on a limited number of studies, the proposed phylogeny represents our best estimate of these relationships and approximate divergence times.
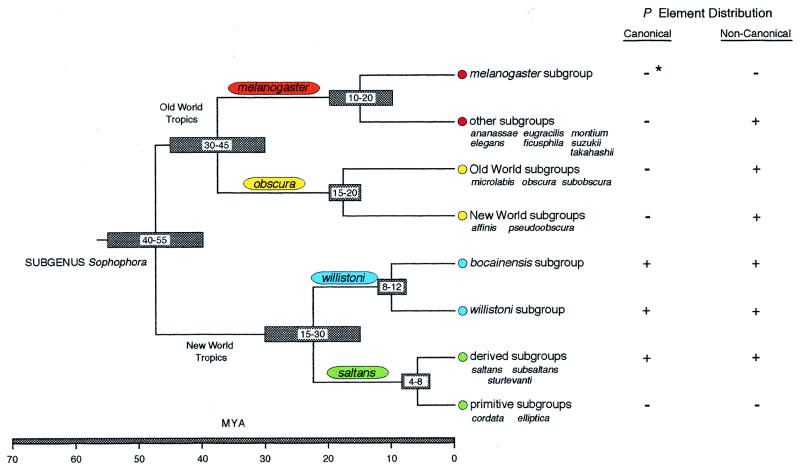
Figure 1.
Phylogeny of the subgenus Sophophora, showing estimates for divergence times and the distribution of both canonical (complete, active elements similar to those in D. melanogaster) and noncanonical P elements. The relationships among the four principal species groups is indicated, and within each species group a major split among subgroups is shown. Phylogeny and divergence times were estimated from a number of morphological, cytogenetic, biogeographical, and molecular studies. Each species group is identified by a particular color, which allows the origin of particular sequences in Fig. 3 to be easily discerned. ∗ in the canonical column for the melanogaster subgroup identifies the P element transferred horizontally to D. melanogaster.
P Element Distribution in Sophophora.
The distribution of P elements in Sophophora was first examined using Southern blots of genomic DNA probed with the canonical P element from D. melanogaster under conditions of reduced stringency (17). This study has been augmented and refined by the PCR survey described here. It is seen that the canonical P elements are confined to the New World saltans and willistoni species groups and, with the exception of the canonical P element in D. melanogaster, are absent from the Old World species groups. This exception is best explained by a previously well documented horizontal transfer of the canonical P element from D. willistoni of the willistoni species group to D. melanogaster (17, 25).
P Element Phylogeny.
To date, P element sequences have been sampled from 40 species, as listed in Table 1. For each species, the PCR was used to amplify genomic copies of the P element. The PCR products for each species were cloned and between four and 14 clones were chosen at random for sequencing. The location of the region flanked by the PCR primers is shown relative to the complete element in Fig. 2. This particular region of the complete P element was chosen for several reasons: (i) The results of a phylogenetic analysis using the limited number of complete P element sequences are consistent with an analysis using this region alone. (ii) The 500-bp region is long enough to yield phylogenetically useful information, but short enough to allow rapid sequencing in a number of species. (iii) Because the divergence of P element sequences in our sample can approach 50%, and because some elements are internally deleted, the choice of primers was limited to certain conserved regions. The primers were designed to preserve amino acid identities in these conserved regions, but codon usage preferences also were considered so that the degeneracy of the primers could be minimized. It is possible that more divergent subfamilies of P elements were not identified with these primer combinations. Thus, the results presented here should be viewed not as an exhaustive survey, but rather as a robust sample of some of the diversity that exists among the P elements in the subgenus Sophophora.
The results of a phylogenetic analysis of consensus P element sequences (see Materials and Methods) are shown in Fig. 3 as a most parsimonious tree chosen at random from 1,000 such trees. P element sequences fall into distinct groups, or subfamilies, designated by letters in Fig. 3. A subfamily is defined as the largest well supported monophyletic group of sequences sampled from a particular species group. (The closely related saltans and willistoni species groups, which share a relatively recent common ancestor, are treated for the purpose of this analysis as a single species group.) In a few cases, single sequences from one species group are found within a P element subfamily of another species group; these exceptions will be discussed below. Clade G is technically not a subfamily because it includes sequences from all four species groups. However, it is a well supported group and is identified to aid the discussion of the results.
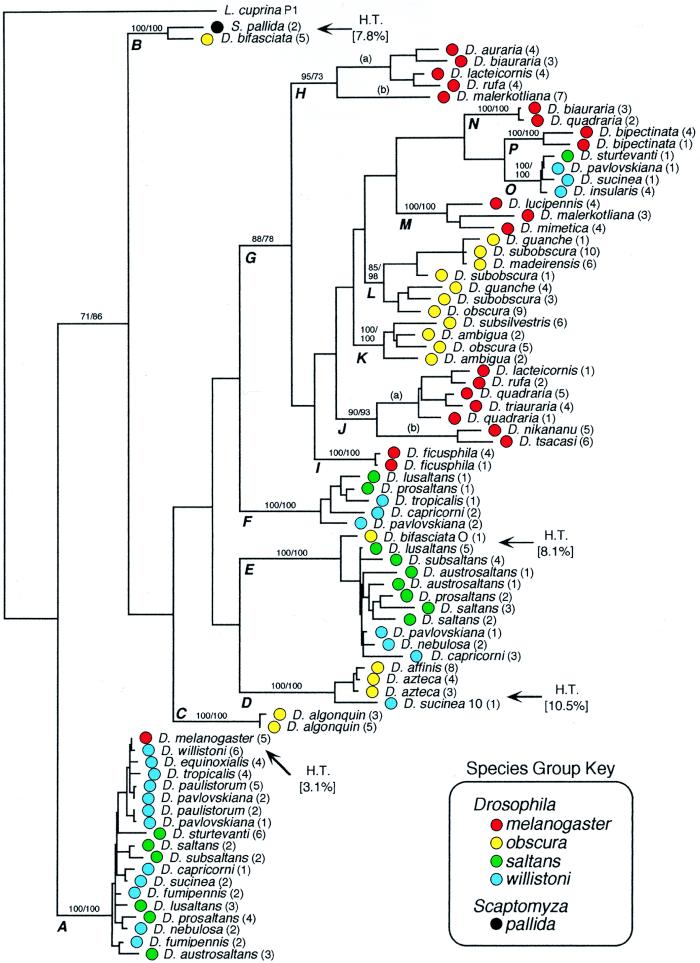
Figure 3.
Phylogenetic analysis of P element nucleotide sequences from the saltans and willistoni species groups. Comparisons were limited to 449 bp between primers 2016 and 2017 and used the sequence from L. cuprina as an outgroup. The data set consists of 80 consensus sequences that were derived from monophyletic sequences obtained from particular species (see Materials and Methods). The total number of sequences used to derive the consensus sequences was 250. Species names are given in italics followed by the total number of clones used to construct the consensus sequence in parentheses. Species group affiliations are indicated by color as shown in the key. This cladogram was generated by parsimony analysis as implemented paup 3.1.1 (23) using the heuristic search algorithm with tree bisection-reconnection branch swapping and random stepwise addition of taxa. This is an arbitrarily chosen representative of 1,000 equally parsimonious trees, each requiring 2,089 steps. The consistency index is 0.402 and the retention index 0.799. Bootstrap analysis of 100 replicates was performed on the data using both parsimony and neighbor-joining. Numbers shown on the branches before the slash are bootstrap percentages derived from parsimony, after the slash from neighbor-joining. Values of 50% or greater are shown only for the major groups. Letters refer to clades that are discussed in the text. Four proposed instances of horizontal transfer (H.T.) are identified by arrows. Within the brackets, the average percent divergence separating the P element sequence from the recipient species and the rest of the sequences in the clade is shown.
P elements from the saltans and willistoni species groups from four subfamilies (clades A, E, F, and O). Clade A represents the canonical P elements that are the most prevalent, comprising over half of the clones sequenced from these two species groups. In spite of being the most numerous P element subfamily sampled, differentiation within this subfamily is less than 10%. This implies that either these sequences are under strong selection to maintain their sequence integrity, or they are relatively recent additions to the saltans-willistoni lineage (see Discussion). Clade E includes about half the number of sequences as clade A, and clades F and O are represented by relatively few sequences. This distribution could reflect the representation of these subfamilies in the genomes of the flies from the saltans and willistoni groups, but probably is influenced to some degree by the sampling done with the PCR primers. Sequences from clade O are noteworthy because they show an affiliation with P elements from both the melanogaster and obscura species groups.
P element sequences from the obscura species group are distributed among four subfamilies, represented by clades C, D, K, and L. The majority of sequences belong to the latter clade, which include P elements only from the Old World obscura subgroup (see Fig. 1). This subfamily includes three sequences, D. subobscura G2 and A1 and D. guanche G1, described previously (14, 15). Clade C comprises eight sequences only from D. algonquin, a member of the affinis subgroup that has an extant distribution in the New World. Clade D consists of 15 sequences from two additional New World species, D. affinis and D. azteca. Also included in clade D is a single sequence from D. sucinea of the willistoni species group. In a previous analysis (13), this sequence, D. sucinea 10, was the only one (of 92 total sequences) to show no affiliation with any of the four saltans-willistoni P element subfamilies. Indeed, the nucleotide sequence divergence between D. sucinea 10 and the rest of the saltans-willistoni sequences ranges from 32% to 37%. As seen in Fig. 3, this sequence differs by an average of only 10.5% from the D. affinis and D. azteca sequences in clade D. It is intriguing that considerable overlap of the geographic distributions of D. sucinea and D. azteca in central Mexico (26) exists.
P elements from D. bifasciata, a widespread species belonging to the Old World obscura subgroup, are unusually interesting. It has been shown previously that this species possesses two distinct subfamilies, called M and O types (18, 19). The former belong to clade B, where they are closely related to P element sequences from Scaptomyza pallida. The O-type sequence belongs to clade E, which is one of four saltans-willistoni subfamilies. It is even more closely related to another P element from S. pallida (27) (not shown in Fig. 3). Thus far, no obscura subgroup P elements (clades K and L) have been identified in D. bifasciata.
Six P element subfamilies are in the melanogaster species group (clades H, I, J, M, N, and P). Clade J includes sequences from six different species, all in the montium subgroup that is the largest in the melanogaster species group. Clade H includes additional sequences from four species of the montium subgroup (a) and from D. malerkotliana of the ananassae subgroup (b). In spite of belonging to the same subfamily, the P elements from the ananassae subgroup are in fact quite distinct (average pairwise divergence of 30%) from the montium sequences. Clade N includes sequences from two closely related species, D. biauraria and D. quadraria. Clade P, comprising five sequences from D. bipectinata, is a sister group to clade O of the saltans and willistoni groups. The average sequence divergence between clades P and O is 16.9% compared with an average sequence divergence between clades P and N of 19.9%. Between clades N and O, the average sequence divergence is 20.4%.
Not apparent from Fig. 3 is the complete absence of detectable P elements in many species of the melanogaster group. An indication is given in Table 1, where detectable P elements are shown to be absent in 10 of 24 species examined (see also ref. 17). As seen in Fig. 1, it is especially interesting that P element sequences are absent in those species most closely related to D. melanogaster. Because they are widely distributed throughout the subgenus Sophophora, the most likely explanation is that the P sequences have been lost from the melanogaster subgroup or have diverged beyond recognition by the primers used here. A more detailed analysis of P element distribution in the melanogaster (J.B.C., P. Kim, and M.G.K., unpublished work) and saltans and willistoni species groups (13) indicates that loss of particular P element subfamilies in certain species has occurred multiple times, suggesting that this is a fairly common occurrence.
On a fine scale, general agreement exists between P element and species phylogenies. For example, clade J comprises P elements from six species of the montium subgroup of the melanogaster species group. Within this clade species-specific branching is found, which reflects the phylogeny of this well studied subgroup. The sequences from the four species of the auraria complex (a), which have an Oriental distribution, are distinct from the sequences from the two African species (b). On a broader scale, however, the P element phylogeny is not congruent with species phylogeny. If the P element phylogeny were to reflect the species phylogeny perfectly, sequences from each of the four species groups would be monophyletic. Furthermore, all sequences from the Old World lineage of Sophophora, represented by the melanogaster and obscura species groups should be monophyletic, as should sequences from the New World lineage, represented by the saltans and willistoni groups. However, the pattern of P element phylogeny in Sophophora is much more complex than this and includes the existence in the same genome of sequences belonging to different subfamilies.
DISCUSSION
P Element Sequence Divergence.
The consensus sequences used here for each species were derived from sequences that show varying degrees of differentiation. For example, divergence among the seven sequences comprising the D. malerkotliana consensus sequence in clade H ranges from 0 to 6.6%. Although identical sequences probably represent the same genomic copy sampled more than once, the most divergent sequence, D. malerkotliana 1N, is clearly a distinct copy because it possesses a unique deletion. Within clade O, the four sequences comprising the D. insularis consensus sequence differ by between 1.7 and 6.1%, whereas nine sequences comprising the D. obscura consensus in clade L differ by between 0.7 and 5.3%. The most likely explanation for P element divergence within a species is sequence differentiation after replicative transposition, the mechanism by which many transposable elements move (28). Although autonomous P elements do not always make copies of themselves when they are mobilized, replicative transposition is achieved in many instances (29). As long as no subsequent exchange occurs between copies (via gene conversion or recombination), they are expected to evolve independently and eventually become differentiated. However, it is clear that sequences from the same species that show only modest sequence divergence have a common evolutionary origin.
In many instances, sequence divergence among P elements from the same species is such that the sequences actually belong to different subfamilies. In contrast to sequences from the same subfamily, these represent sequences that clearly have distinct evolutionary histories. For example, sequences from D. pavlovskiana are represented in each of the four saltans-willistoni subfamilies shown in Fig. 3 (clades A, E, F, and O). The pairwise sequence divergence among these sequences is between 30% and 34%. Two explanations could account for the existence of these and other P element subfamilies. First, they could represent ancestral polymorphisms that arose before the divergence of the species themselves. If this is true, then some phylogenetic comparisons may, in fact, include sequences that are in a sense paralogous, explaining some aspects of the lack of congruence between P element and species phylogenies. Second, they could represent sequences that were introduced into particular Sophophora lineages at various times by horizontal transfer. It should be stressed that these two explanations are not mutually exclusive, and each may have been involved in forming the pattern of the phylogeny revealed by this study.
Horizontal Transfer.
Because clade G is the only clade with P elements from all four major Sophophora species groups, it could represent extant members of an ancestral P element that was present before the diversification of the species groups. Other P element subfamilies, that lie outside of clade G, could represent additional, ancestral subfamilies that were retained in some species but not in others. However, it is intriguing that all of the Drosophila sequences that lie outside of clade G were isolated from species that are either relatively widespread (D. bifasciata), cosmopolitan (D. melanogaster), or are found only in the New World (D. affinis, D. azteca, D. algonquin, and all members of the saltans and willistoni species groups). This raises the possibility that these sequences (clades A, B, C, D, E, and F) were introduced into these species by horizontal transfer from donor species with New World or cosmopolitan distributions. This scenario seems very likely for the M-type elements of D. bifasciata (clade B), where the donor species, from the genus Scaptomyza, actually has been identified. What’s more, the P elements from the cosmopolitan species S. pallida are known to be active when transferred to other species in the laboratory (20).
Given their position toward the root of the phylogenetic tree in Fig. 3, the possibility of an origin outside of the subgenus Sophophora is quite strong for the canonical P elements in clade A. If this is true, then the origin of the canonical P elements can be dated to somewhere between 15–30 million years ago (the divergence of the two New World subgroups) and 40–55 million years ago (the divergence of the Old World and New World subgroups). This would explain why, with one exception, they are confined to the New World saltans and willistoni species groups and perhaps explain their relatively modest sequence differentiation compared with other sequences in the saltans and willistoni species groups (13). Although these observations constitute circumstantial evidence, this hypothesis cannot be substantiated until a donor species is identified.
In four instances, horizontal transfer is clearly the most likely explanation for lack of congruence between species phylogeny and P element phylogeny (see Fig. 3, arrows). The first two of these cases of horizontal transfer have been previously identified (see ref. 25): (i) The identity of the P elements from D. melanogaster of the melanogaster species group to that of D. willistoni of the willistoni species group (clade A). (ii) The affiliation of the M-type P elements from D. bifasciata of the obscura species group with sequences from S. pallida (clade B). Two additional cases of horizontal transfer are now apparent, demonstrating the power of this combined phylogenetic analysis. (iii) The affiliation of the O-type P element from D. bifasciata with those of the saltans-willistoni subfamily in clade E. (iv) The presence of D. sucinea 10 (willistoni species group) within clade D, which includes sequences from two New World species of the obscura group. Together these represent four instances of horizontal transfer, apparent because of the marked discrepancy between P element and host phylogenies. Alternative explanations for these discrepancies can be considered, but they require certain assumptions that are not as plausible as those for horizontal transfer (25).
Compelling evidence shows that some P element lineages may have been involved in more than one horizontal transfer. For example, the canonical P element (clade A) may have been involved in at least two separate transfers, one from an unknown donor to the saltans-willistoni lineage and a second from D. willistoni to D. melanogaster. The fact that the P element phylogeny within clade A does not always trace species phylogeny (see Fig. 3 and ref. 13) suggests that additional transfers of this active element may have occurred between some species in the saltans and willistoni groups. With respect to multiple horizontal transfers, the O-type P element from D. bifasciata (clade E) is particularly interesting. Whereas this sequence differs from those of the saltans and willistoni groups by an average of 8.1%, it differs by less than 1% from another P element isolated from S. pallida (27) (the region compared in that study does not overlap the PCR fragment used here and hence the O-type sequence from S. pallida is not included in the analysis presented in Fig. 3). Thus, this element may have been involved in three separate horizontal transfers: from an unknown donor to the saltans-willistoni lineage, from a species in this lineage to S. pallida, and from S. pallida to D. bifasciata. This scenario seems more likely than transfer from saltans-willistoni to D. bifasciata and then to S. pallida because S. pallida is a cosmopolitan species, which could span the distribution of both the New World saltans and willistoni groups and the Old World species D. bifasciata.
Several properties of the biology of active P elements are consistent with the hypothesis of horizontal transfer. First, they possess all of the properties necessary for mobility in the appropriate cytological background. This includes encoding the transposase necessary for mobility. Second, both theoretical (30) and laboratory studies (31) demonstrate that the high transposition efficiency of P elements in naive genomes overrides any deleterious effects associated with transposition. Third, during transposition the P element usually (but not always) makes a copy of itself (32). This replicative mode of transposition leads to an increase in genomic P element copy number over time. Given these properties, if an active P element is introduced into the genome of a naive species that could support transposition, the chances seem reasonably good that it would spread throughout the population. Although the exact mechanism of horizontal transfer is unknown, the minimum requirement would be geographic, temporal, and ecological overlap between donor and recipient species. D. willistoni and D. melanogaster have become sympatric species only through the activities of modern human travel and commerce, probably within the past 200 years (33). It is not known exactly when the actual horizontal transfer of the canonical P element from D. willistoni to D. melanogaster occurred, but it is known that the P element has spread throughout worldwide populations of the former species only within the past 30–40 years (34, 35). Furthermore, the DNA sequence identity of P elements from diverse geographic locations is consistent with a very recent introduction into D. melanogaster.
Compared with nonmobile nuclear genes, transposable elements exhibit a propensity for horizontal transfer (see ref. 10). This is undoubtedly due to those properties, such as replicative transposition, discussed above. For the mariner transposable element, which like P is a Class II transposable element, horizontal transfer occurs relatively frequently and even between distant taxa (36, 37). Furthermore, it has been suggested that horizontal transfer may, in fact, be essential to the long-term survival of mariner and other transposable elements (12). In the absence of horizontal transfer to a new host species, the fate of transposable elements may be sequence degeneration and eventual loss from the genome through stochastic processes (38). Thus, it can be argued that the persistence of transposable elements, like mariner and P, is indirect evidence for the phenomenon of horizontal transfer.
The implications of this study extend beyond the evolution of P elements in Drosophila. First, transposable elements are ubiquitous features of eukaryotes and in some species comprise a significant portion of the genome (39, 40). Thus, information on the evolution of transposable elements is important for understanding the evolution of eukaryotic genomes as a whole. This takes on significance with increasing evidence that transposable elements may play integral roles in gene regulation (41, 42) and the generation of host genetic variability (43). Second, the study of transposable element evolution has led to the identification of biological phenomena, such as horizontal transfer, that seem to be extremely infrequent in studies of nonmobile genes. Third, transposable elements present special challenges to phylogenetic biology because of their multicopy nature and their propensity for horizontal transfer. Studies of transposable elements may help to identify potential problems with phylogenetic analysis that are associated with these characteristics and in the process provide insights into the practice of molecular phylogenetics. Finally, knowledge of the evolution of transposable elements may provide insights into the potential opportunities for the population spread of genetically engineered genes and the constraints on such spread that may occur within an evolutionary context.
Table 1.
Distribution of P element sequences among the four principal species groups of the subgenus Sophophora
| Species group | Number of subgroups examined (total number) | Number of species with detectable P sequences (number of species examined) | Number of clones sequenced |
|---|---|---|---|
| melanogaster | 8 (11) | 14 (24) | 76 |
| obscura | 2 (4) | 10 (14) | 73 |
| saltans | 5 (5) | 6 (8) | 41 |
| willistoni | 2 (2) | 10 (10) | 49 |
| Total | 17 (22) | 40 (56) | 239 |
Acknowledgments
We thank Rosa de Frutos and her colleagues at the University of Valencia (Spain) for providing some of the sequence data from the obscura species group. We are grateful to Joana Silva and C. William Birky, Jr. for comments on the manuscript, to Joana Silva for discussions on P element evolution, and to Bill Heed for discussions of Drosophila systematics and evolution. This work was supported by National Science Foundation Grant DEB-9119349.
References
- 1.Finnegan D J. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 2.Kidwell M G, Kidwell J F, Sved J A. Genetics. 1977;36:813–33. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Hare K, Rubin G M. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- 4.Engels W R. In: Mobile DNA. Berg D, Howe M, editors; Berg D, Howe M, editors. Washington, D.C.: Am. Soc. Microbiol.; 1989. pp. 437–484. [Google Scholar]
- 5.Eanes W F, Wesley C S, Hey J, Houle D, Ajioka J W. Genet Res. 1988;52:17–26. [Google Scholar]
- 6.Fitzpatrick B J, Sved J A. Genet Res. 1986;48:89–94. [Google Scholar]
- 7.Mackay T F C. BioEssays. 1996;18:113–121. doi: 10.1002/bies.950180207. [DOI] [PubMed] [Google Scholar]
- 8.Kidwell M G. J Hered. 1994;85:339–346. doi: 10.1093/oxfordjournals.jhered.a111478. [DOI] [PubMed] [Google Scholar]
- 9.Preston C R, Engels W R. Prog Nucleic Acid Res Mol Biol. 1989;36:71–85. doi: 10.1016/s0079-6603(08)60162-2. [DOI] [PubMed] [Google Scholar]
- 10.Kidwell M G. Genetica. 1992;19:913–916. doi: 10.1007/BF00133726. [DOI] [PubMed] [Google Scholar]
- 11.Kidwell M G. Annu Rev Genet. 1993;27:235–256. doi: 10.1146/annurev.ge.27.120193.001315. [DOI] [PubMed] [Google Scholar]
- 12.Lohe A R, Moriyama E N, Lidholm D A, Hartl D L. Mol Biol Evol. 1995;12:62–72. doi: 10.1093/oxfordjournals.molbev.a040191. [DOI] [PubMed] [Google Scholar]
- 13.Clark J B, Altheide T K, Schlosser M J, Kidwell M G. Mol Biol Evol. 1995;12:902–913. doi: 10.1093/oxfordjournals.molbev.a040267. [DOI] [PubMed] [Google Scholar]
- 14.Paricio N, M, Perez-Alonso M, Martínez-Sebastián M J, de Frutos R. Nucleic Acids Res. 1991;19:6713–6718. doi: 10.1093/nar/19.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller W J, Hagemann S, Reiter E, Pinsker W. Proc Natl Acad Sci USA. 1992;89:4018–4022. doi: 10.1073/pnas.89.9.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lansman R A, Shade R O, Grigliatti T A, Brock H W. Proc Natl Acad Sci USA. 1987;84:6491–6495. doi: 10.1073/pnas.84.18.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels S B, Peterson K R, Strausbaugh L D, Kidwell M G, Chovnick A. Genetics. 1990;124:339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagemann S, Miller W J, Pinsker W. Mol Gen Genet. 1994;244:168–175. doi: 10.1007/BF00283519. [DOI] [PubMed] [Google Scholar]
- 19.Hagemann S, Miller W J, Pinsker W. Nucleic Acids Res. 1992;20:409–413. doi: 10.1093/nar/20.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonelig M, Anxolabéhère D. Proc Natl Acad Sci USA. 1991;88:6102–6106. doi: 10.1073/pnas.88.14.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins H D, Howells A J. Proc Natl Acad Sci USA. 1992;89:10753–10757. doi: 10.1073/pnas.89.22.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maddison W P, Maddison D. macclade: Analysis of Phylogeny and Character Evolution, Version 3.01. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 23.Swofford D. paup: Phylogenetic Analysis Using Parsimony, Version 3.1.1. Washington, D.C.: Smithsonian Institution; 1993. [Google Scholar]
- 24.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Clark J B, Maddison W P, Kidwell M G. Mol Biol Evol. 1994;11:40–50. doi: 10.1093/oxfordjournals.molbev.a040091. [DOI] [PubMed] [Google Scholar]
- 26.Patterson J T, Mainland G B. Univ Texas Publ. 1944;4445:9–101. [Google Scholar]
- 27.Hagemann S, Haring E, Pinsker W. Genetica. 1996;98:43–51. doi: 10.1007/BF00120217. [DOI] [PubMed] [Google Scholar]
- 28.Berg D E, Howe M M. In: Mobile DNA. Berg D, Howe M, editors; Berg D, Howe M, editors. Washington, D.C.: Am. Soc. Microbiol.; 1989. , 972 pp. [Google Scholar]
- 29.Gloor G B, Nassif N A, Johnson Schlitz D M, Preston C R, Engels W R. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 30.Uyenoyama M, Nei M. Theor Popul Biol. 1985;27:176–201. doi: 10.1016/0040-5809(85)90009-7. [DOI] [PubMed] [Google Scholar]
- 31.Kiyasu P K, Kidwell M G. Genet Res. 1984;44:251–259. doi: 10.1017/s0016672300026495. [DOI] [PubMed] [Google Scholar]
- 32.Engels W R, Johnson-Schlitz D M, Eggleston W B, Sved J. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 33.Engels W R. BioEssays. 1992;14:681–686. doi: 10.1002/bies.950141007. [DOI] [PubMed] [Google Scholar]
- 34.Kidwell M G. Proc Natl Acad Sci USA. 1983;80:1655–1659. doi: 10.1073/pnas.80.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anxolabéhère D, Kidwell M G, Periquet G. Mol Biol Evol. 1988;5:252–269. doi: 10.1093/oxfordjournals.molbev.a040491. [DOI] [PubMed] [Google Scholar]
- 36.Robertson H M, Lampe D J. Annu Rev Entomol. 1995;40:333–357. doi: 10.1146/annurev.en.40.010195.002001. [DOI] [PubMed] [Google Scholar]
- 37.Robertson H M, MacLeod E G. Insect Mol Biol. 1993;2:125–139. doi: 10.1111/j.1365-2583.1993.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan N, Darden T, Langley C. Genetics. 1985;109:459–480. doi: 10.1093/genetics/109.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggesse C, Gatti M. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 41.Britten, R. J. (1997) Gene, in press.
- 42.Britten R J. Mol Phylogenet Evol. 1996;5:13–17. doi: 10.1006/mpev.1996.0003. [DOI] [PubMed] [Google Scholar]
- 43.Kidwell M G, Lisch D. Proc Natl Acad Sci USA. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]