Abstract
Cytochrome P450s constitute a superfamily of genes encoding mostly microsomal hemoproteins that play a dominant role in the metabolism of a wide variety of both endogenous and foreign compounds. In insects, xenobiotic metabolism (i.e., metabolism of insecticides and toxic natural plant compounds) is known to involve members of the CYP6 family of cytochrome P450s. Use of a 3′ RACE (rapid amplification of cDNA ends) strategy with a degenerate primer based on the conserved cytochrome P450 heme-binding decapeptide loop resulted in the amplification of four cDNA sequences representing another family of cytochrome P450 genes (CYP28) from two species of isoquinoline alkaloid-resistant Drosophila and the cosmopolitan species Drosophila hydei. The CYP28 family forms a monophyletic clade with strong regional homologies to the vertebrate CYP3 family and the insect CYP6 family (both of which are involved in xenobiotic metabolism) and to the insect CYP9 family (of unknown function). Induction of mRNA levels for three of the CYP28 cytochrome P450s by toxic host-plant allelochemicals (up to 11.5-fold) and phenobarbital (up to 49-fold) corroborates previous in vitro metabolism studies and suggests a potentially important role for the CYP28 family in determining patterns of insect–host-plant relationships through xenobiotic detoxification.
Keywords: xenobiotic induction, insect–plant interactions, Drosophila, cDNA cloning
Cytochrome P450s constitute a superfamily of heme-thiolate proteins characterized by a highly conserved FXXGXXXCXG sequence of amino acids (heme-binding decapeptide) and spectral absorbance peak at 450 nm (1). The diversity of cytochrome P450s is well established, in terms of both the reactions they catalyze and the chemically dissimilar substrates upon which they act. Functions include the oxidative, peroxidative, and reductive metabolism of steroids, fatty acids, pharmaceuticals, agrochemicals, and plant allelochemicals. Excellent reviews of this superfamily may be found in Nelson et al. (2) and Feyereisen (3).
Cytochrome P450s are involved in insect growth and development through the processing of such compounds as pheromones and ecdysteroids (4, 5). They have also been implicated in metabolic resistance to insecticides, including carbamates, chlorinated hydrocarbons, organophosphates, and pyrethroids (6, 7). Similarly, cytochrome P450-catalyzed elimination of toxic plant allelochemicals appears to be a key factor in host-plant utilization (8, 9).
The most extensively studied group of insect cytochrome P450s is the CYP6 family, which is related to the major drug-metabolizing CYP3 family in vertebrates. In the house fly (Musca domestica), CYP6A1 metabolizes the insecticides aldrin and heptachlor (10), and CYP6D1 has been linked to deltamethrin metabolism (11). Heterologous expression of Drosophila melanogaster CYP6A2 in Saccharomyces cerevisiae bioactivates some genotoxins (e.g., aflatoxin B1) (12). This broad catalytic diversity is thought to have arisen as a result of coevolution between herbivorous animals and toxic allelochemical-producing plants (13). Although this hypothesis is attractive, there have been very few studies that have examined the metabolism of natural substrates by individual cytochrome P450 isoforms. To date, only the CYP6B genes of papilionid caterpillars have been shown to metabolize a natural substrate—toxic furanocoumarins present in the organisms’ host plants (14).
The well-defined interrelationships between columnar cacti and Drosophila in the Sonoran Desert of the southwestern United States and northwestern Mexico provide an ideal model system with which to investigate cytochrome P450-mediated resistance to toxic plant allelochemicals. Four species of Drosophila (i.e., D. mettleri, D. nigrospiracula, D. mojavensis, and D. pachea) utilize, as a feeding and breeding substrate, the necrotic tissue of five species of columnar cacti—i.e., saguaro (Carnegiea gigantea), cardón (Pachycereus pringlei), senita (Lophocereus schottii), agria (Stenocereus gummosus), and organ pipe (Stenocereus thurberi). Allowing for geographic differences in host-plant availability, there is essentially a one-to-one relationship between each drosophilid species and the species of host cactus utilized. Furthermore, while the cactophilic drosophilids are endemic to the Sonoran Desert, they are not phylogenetically close and, therefore, are believed to have evolved independently into the desert niche (15).
Each species of cactus is characterized by a suite of allelochemicals that are toxic to all but normal resident species. In agria and organ pipe cacti (which are utilized only by D. mojavensis), triterpene glycosides, C8–C12 fatty acids, and sterol diols are the major toxins. For D. nigrospiracula (which lives only on saguaro and cardón), D. mettleri (which lives in rot exudate-soaked soils usually found at the base of saguaro and cardón), and D. pachea (which lives exclusively on senita), utilization involves resistance to isoquinoline alkaloids. In saguaro, the alkaloids gigantine and carnegine constitute 1–2% of the plant’s dry weight, whereas in senita, lophocereine and its trimer, pilocereine, constitute up to 20% of dry tissue weight (16). In contrast to the Sonoran Desert species, Drosophila hydei is a cosmopolitan species known to utilize Opuntia cactus tissue, which lacks toxic allelochemicals.
The successful utilization of cactus tissue requires the ability to tolerate these toxins. Several studies have established the involvement of cytochrome P450s in cactophilic drosophilid resistance to isoquinoline alkaloids. In particular, in vitro alkaloid metabolism and total cytochrome P450 content were significantly induced by exposure of desert drosophilids to cactus alkaloids or phenobarbital (an inducer of many toxin-metabolizing cytochrome P450s). Conversely, the induced in vitro metabolism and in vivo larval viability were reduced by the cytochrome P450 inhibitor piperonyl butoxide (8, 17).
Among the cactophilic Drosophila, only D. mettleri displays a behavioral preference for oviposition in rot exudate-soaked soils. As a result of inevitable evaporative water loss from these soils, D. mettleri larvae regularly encounter concentrations of toxic alkaloids that may be more than an order of magnitude higher than in necrotic tissue (18). No other cactophilic drosophilid is routinely exposed to such high levels of toxic plant allelochemicals. This is especially true for larvae in soils soaked by rot exudate from senita cactus. In the current study, therefore, the induction of individual D. mettleri cytochrome P450s by senita alkaloids was investigated. We have identified a previously undescribed family of insect P450s as being potentially involved in the metabolism of these host-plant compounds.
MATERIALS AND METHODS
Drosophila Species and Larval Induction.
Multifemale lines of D. mettleri and D. nigrospiracula originally collected from mainland regions of the Sonoran Desert and D. hydei (strain A820) originally isolated from Jalisco, Mexico, were used in the current study. Drosophila stocks were maintained on yeast-supplemented instant Drosophila medium (Ward’s, Rochester, NY). For the initial cytochrome P450 cDNA cloning work, early third-instar larval Drosophila were induced for 48 hr prior to RNA isolation with 20 mg of phenobarbital dissolved in 1 ml of water (2% wt/vol, pH 9.0) and distributed evenly across the surface of 50 g of rehydrated instant Drosophila medium. As an inducer of several cytochrome P450s implicated in xenobiotic metabolism, treatment with phenobarbital was used to maximize the message levels of genes potentially involved in metabolism of toxic isoquinoline alkaloids. To obtain RNA for use in Northern analyses, early third-instar larvae were induced for 48 hr with 1 g (dry weight) of one of the following: (i) senita cactus tissue containing lophocereine and pilocereine at concentrations present in fresh tissue (i.e., 15% dry weight), (ii) saguaro tissue supplemented with purified saguaro alkaloids to a concentration of 3 or 10 times that found in fresh tissue (i.e., 4.5% or 15% dry weight), (iii) purified agria triterpene glycosides (≈3 times the concentration present in fresh tissue), or (iv) 20 mg of phenobarbital, as described above. All inducers were distributed evenly across the surface of 50 g of instant Drosophila medium and rehydrated before the introduction of 1 g of larvae. Uninduced larvae maintained on instant Drosophila medium served as controls. Saguaro alkaloids and agria triterpene glycosides were extracted from dried cactus tissue by using standard methods (18, 19).
RNA Isolation and cDNA Synthesis.
Total RNA for the initial 3′ rapid amplification of cDNA ends (RACE) (20) reaction was isolated from phenobarbital-induced larvae by following the guanidine⋅hydrochloride-based method of Skuse and Sullivan (21). Poly(A)+ RNA was isolated for 5′ RACE reactions (20) and Northern analyses by direct capture of mRNA onto oligo(dT)25-coated paramagnetic beads (Novagen), following the procedure of Jakobsen et al. (22).
Because full-length cDNAs are not essential for cloning of the 3′ ends of cytochrome P450 cDNAs, total RNA and standard Moloney murine leukemia virus reverse transcriptase were used for first-strand synthesis primed off the poly(A) tail. Where a high percentage of full-length cDNAs was required (e.g., cloning of cytochrome P450 5′ sequences), Superscript II (GIBCO/BRL), which lacks RNase H activity, was employed for first-strand cDNA synthesis.
Cytochrome P450 cDNA Cloning.
For the 3′ RACE reaction, a fully degenerate (2,048-fold) gene-specific primer [5′-GGICCI(A/C)GIAA(C/T)TG(C/T)ATIGC-3′, where I represents deoxyinosine and the parentheses contain mixed bases] was designed on the basis of the highly conserved GPRNCIG heme-binding decapeptide motif common to many insect cytochrome P450s. Deoxyinosine was used at sites of fourfold degeneracy to minimize mismatch instability. Using sequence information obtained from the 3′ end of PCR-amplified cytochrome P450 cDNAs, gene-specific reverse primers (CYP28A1, 5′-CCGAGCGAGTCTTTGGATTGA-3′; CYP28A2, 5′-CCTGGCATTGACACTGACATC-3′) were constructed and full-length cDNAs were obtained by 5′ RACE. Consensus cDNA sequences were constructed with data from multiple overlapping clones to minimize errors due to nucleotide misincorporation by reverse transcriptase and Taq DNA polymerase.
rp49 cDNA Cloning.
Partial cDNA sequence for ribosomal protein 49 (rp49) (homologous to human ribosomal protein L32) was cloned by 3′ RACE from each of the Drosophila species for use as a homologous probe to standardize mRNA loading in the Northern analyses. The sequence of the rp49 gene-specific primer was 5′-AA(A/G)TTC(C/T)TGGTGCA(C/T)AA(C/T)GT-3′. Ribosomal proteins are highly conserved genes that display stable high-level expression (23). The small size of the rp49 transcript (approximately 650 bp) made its use as a loading standard ideal because its signal appeared well below that of the cytochrome P450 transcripts.
DNA Sequencing and Analysis.
Amplification products were ligated into the pGem-T vector (Promega) and used to transform Escherichia coli strain DH5-α. Inserts from transformants were sequenced by the dideoxynucleotide chain-termination method (24) and analyzed for similarity to known genes with the blast algorithm at the National Center for Biotechnology Information (25). Using the blast-generated regional sequence alignments as a guide, we manually aligned the deduced full-length amino acid sequences for CYP28A1 and CYP28A2 with the sequences for D. melanogaster CYP6A2 (GenBank M88009) and CYP4D1 (GenBank X67645), house fly, Musca domestica, CYP6A1 (GenBank L27241) and CYP6D1 (GenBank U22362), black swallowtail butterfly, Papilio polyxenes, CYP6B1 (GenBank M80828) and CYP6B3 (GenBank U25819), Australian cotton bollworm, Helicoverpa armigera, CYP6B2 (GenBank U18085), tobacco budworm, Heliothis virescens, CYP9A1 (GenBank U23506), cockroach, Blaberus discoidalis, CYP4C1 (GenBank M63798), rat, Rattus norvegicus, CYP3A1 (GenBank D29967), and the human CYP3A4 (GenBank J04449) and CYP11A1 (GenBank D00169). Distance matrices and phylograms were generated from aligned sequences by using paup software (Phylogenetic Analysis Using Parsimony, by D. Swofford, Smithsonian Institution). Human CYP11A1 served as the outgroup for the phylogenetic analyses.
Northern Analyses.
Induction of individual cytochrome P450s by alkaloid-containing cactus tissue, agria triterpene glycosides, or phenobarbital was investigated by high-stringency Northern hybridization using poly(A)+ RNA blotted onto Hybond N+ nylon membrane (Amersham) and digoxigenin-UTP-labeled probes. Detection of bound probe employed anti-digoxigenin Fab conjugated to alkaline phosphatase, which catalyzed the degradation of the chemiluminescent substrate CDP-Star (Tropix, Bedford, MA), following the protocol of Engler-Blum et al. (26). Photographic images (X-Omat AR film, Kodak) of the chemiluminescent signal for each cytochrome P450 and a subsequent probing of the same blot for rp49 transcripts were superimposed, scanned at 300 dots per inch, and analyzed using Photoshop software. Signal strength was quantified on the basis of the above-background pixel counts of the scanned image and standardized to signal from a second probing of message for rp49 based on the linear (for the blots analyzed) relationship between the amount of RNA loaded and the resulting signal intensity.
RESULTS
Sequencing and blast analysis of 123 subcloned cDNAs amplified by 3′ RACE using a moderately degenerate oligonucleotide primer targeted to sequence encoding the heme-binding region of insect cytochrome P450s resulted in the identification of 101 partial cytochrome P450 cDNAs. These sequences represented 15 distinct forms of cytochrome P450, which displayed strong regional similarity to members of the CYP6 (5 forms), CYP4 (6 forms), and CYP9 (2 forms) cytochrome P450 families.
Each of the 15 partial cDNA sequences from D. mettleri was used as a probe to screen the corresponding cytochrome P450 transcripts (by Northern hybridization) for xenobiotic responsiveness to senita cactus alkaloids. Only three cytochrome P450 genes (initially designated DU43, DU108, and DU369) demonstrated a greater than 3-fold increase in message following exposure to senita alkaloids. In accordance with established cytochrome P450 nomenclature, DU369 and DU43 have been designated CYP28A1 (GenBank U89746) and CYP28A2 (GenBank U89747), respectively (DU108 has not been assigned a CYP designation).
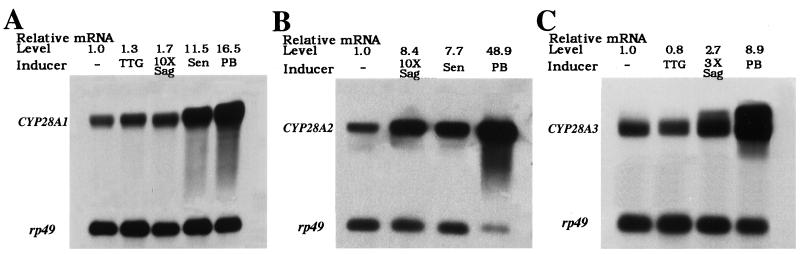
Northern blots for the CYP28 transcripts are shown in Fig. 1. While CYP28A2 (Fig. 1B) and DU108 were induced to similar degrees following exposure to senita cactus alkaloids (7.7-fold and 7.1-fold, respectively), CYP28A1 displayed greater than 11-fold induction (Fig. 1A). Interestingly, xenobiotic induction of CYP28A1 was specific for senita alkaloids, as neither chemically similar saguaro alkaloids (at an equivalent concentration) nor toxic agria cactus triterpene glycosides were able to induce a similar response. By contrast, CYP28A2 and DU108 were crossinducible by saguaro cactus alkaloids (8.4-fold and 9.3-fold, respectively) but not by agria triterpene glycosides. Finally, all three genes showed a greater than 10-fold increase in message with exposure to phenobarbital (CYP28A2, 48.9-fold; DU108, 26-fold; and CYP28A1, 16.5-fold).
Figure 1.
Analysis by Northern hybridization of the responsiveness of CYP28A1 (A), CYP28A2 (B), and CYP28A3 (C) to agria cactus triterpene glycosides (TTG), saguaro cactus alkaloids (Sag), senita cactus alkaloids (Sen), and phenobarbital (PB). Induction relative to untreated controls (−) was standardized to signal from a homologous rp49 probe. Images of the sequential cytochrome P450 and rp49 probings of the same blot have been superimposed. (Note: Interference from residual rRNA produces the “doublet-like” bands in the 3× Sag and PB lanes of C.)
Because of the magnitude and specificity of alkaloid responsiveness displayed by CYP28A1, the partial cDNA sequence was used to design a gene-specific reverse primer and the remaining cDNA was obtained by 5′ RACE. Full-length cDNA sequence was also obtained for CYP28A2, the cytochrome P450 most closely related to CYP28A1. The consensus nucleotide sequences and conceptual amino acid translations of the full-length cDNAs obtained are shown in Fig. 2 A and B. The 1,831-bp cDNA for clone CYP28A1 and the 1,764-bp cDNA for clone CYP28A2 contain open reading frames encoding proteins of 506 and 505 residues, respectively. The 3′ untranslated regions of CYP28A1 and CYP28A2 both contain a putative polyadenylation signal [AATAAT and AATAAA, respectively (27)] within 15 bases of the poly(A) tail.
Figure 2.
Nucleotide and predicted amino acid sequences of CYP28A1 (A) and CYP28A2 (B). Amino acids are numbered on the left and nucleotides, on the right. Regions of conservation associated with helix I, helix K, and the heme-binding decapeptide are single-underlined. Putative polyadenylation signals are double-underlined.
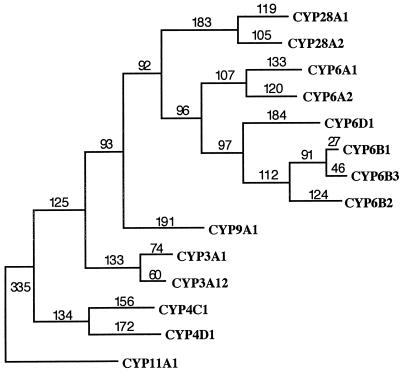
Alignment of the deduced amino acid sequence for both CYP28A1 and CYP28A2 by blast revealed strong regional similarity (up to 64%) to several CYP3A and CYP6A sequences for the region encompassing the C terminus of helix I through the middle of helix L (CYP28A1 residues G308–F471 and CYP28A2 residues G307–F470). The heme-binding decapeptide is located just N-terminal to helix L. Total positional identity for full-length CYP28 sequences aligned with previously described cytochrome P450s, however, did not exceed 25%. By contrast, an alignment between CYP28A1 and CYP28A2 revealed 55.6% sequence identity. The relatedness of the CYP28 sequences to each other and their phylogenetic uniqueness among insect cytochrome P450s is also evident in the single, most parsimonious, phylogenetic tree generated by heuristic analysis of full-length protein sequences (Fig. 3). The CYP28 sequences form a monophyletic clade that is separate from the clades formed by members of the CYP3, CYP6, and CYP9 families. Both CYP3 and CYP6 have been implicated in xenobiotic metabolism. The function of CYP9 has not been determined. While a phylogenetic tree based on maximum parsimony suggests that the CYP6 and CYP28 families may have been derived from a common ancestor, bootstrap analysis produced a polytomy, with a bootstrap value of 69, among the monophyletic clades for the CYP3, CYP6, CYP9, and CYP28 sequences.
Figure 3.
Phylogram based on the single most parsimonious tree generated from the aligned full-length amino acid sequences of CYP28A1, CYP28A2, and representatives of the CYP3, CYP4, CYP6, and CYP9 families. The human mitochondrial CYP11A1 served as the outgroup. Branch lengths are indicated in terms of the predicted number of amino acid changes.
Sequence similarity was used to identify additional members of the CYP28 family from among 3′ RACE-generated partial cytochrome P450 cDNAs for two other drosophilid species, D. nigrospiracula and D. hydei. An alignment of the deduced amino acid sequences encoded by these genes with the corresponding region (helix L through the translational stop codon) of CYP28A1 and CYP28A2 is presented in Fig. 4. In pairwise comparisons of clone DU33ng (the most closely related sequence from D. nigrospiracula) and clone DU4hy (the most closely related sequence from D. hydei) with CYP28A1, amino acid positional identity was 72% and 81%, respectively. Compared with CYP28A2, identity dropped to 56% in both cases. This was the same level of identity seen between CYP28A1 and CYP28A2. The sequences for DU33ng and DU4hy have been deposited in GenBank and designated CYP28A3 (accession no. U91565) and CYP28A4 (accession no. U91566), respectively.
Figure 4.
Alignment of the deduced amino acid sequences (helix L through translational stop) of CYP28A3 and CYP28A4 with CYP28A1 and CYP28A2. Identical amino acids are indicated by periods and differences, by the appropriate amino acid single-letter symbol.
Northern analyses of CYP28A3 from D. nigrospiracula to test for induction by phenobarbital and cactus allelochemicals are shown in Fig. 1C. There was an 8.7-fold increase in message with phenobarbital and a 2.7-fold increase with saguaro alkaloids at a concentration approximately 3 times higher than normally found in fresh cactus tissue. No induction was seen after exposure to the toxic triterpene glycosides of agria cactus, which cannot be utilized by D. nigrospiracula as a host plant. The toxicity of senita alkaloids and of higher concentrations of saguaro alkaloids to D. nigrospiracula precluded their use in tests for induction. For the same reason, it was impossible to screen CYP28A4 from the noncactophilic D. hydei for responsiveness to any toxic cactus allelochemicals.
DISCUSSION
The genetic and functional diversity of the cytochrome P450 superfamily is well established, particularly in mammals. A suite of molecularly driven DNA turnover events including nonhomologous crossover, gene amplification, gene conversion, transposition, and replication slippage is thought to play a central role in the expansion and evolution of the cytochrome P450 superfamily. While the first cytochrome P450 may have originated 3.5 billion years ago, there has been a dramatic increase in the number of new cytochrome P450s within the past 400 million years. This, it has been postulated, reflects coevolution between plants that produce toxic secondary compounds to deter herbivory and the enzymatic detoxification systems of animals that feed on these plants (2, 13). Few studies, however, have looked directly at the role of individual cytochrome P450 isoforms in the metabolism of plant allelochemicals. The well-defined insect–host-plant relationships of Sonoran Desert Drosophila, in general, and the resistance of D. mettleri to the toxic isoquinoline alkaloids of the senita cactus, in particular, have provided an ideal context in which to investigate this issue. Furthermore, the use of PCR-based cloning strategies has greatly facilitated the isolation of numerous distantly related cytochrome P450 genes. This is an essential first step for any comprehensive investigation into the relative metabolic contributions of different cytochrome P450 isoforms.
The first indication that specific cytochrome P450 genes may encode xenobiotic-metabolizing enzymes can be seen in their responsiveness at the mRNA level to xenobiotic exposure. In contrast to most housekeeping genes, many xenobiotic-metabolizing cytochrome P450s are strongly inducible by substrates upon which they act (28). Given the relatively close phylogenetic relationship between the xenobiotic-inducible vertebrate CYP3 cytochrome P450s, which figure prominently in drug metabolism (29), and invertebrate CYP6 sequences, it is not surprising that several of these genes have also been implicated in the metabolism of insecticides and toxic plant allelochemicals.
Northern hybridization of mRNA from third-instar D. mettleri larvae that had been previously exposed to senita alkaloids (lophocereine and pilocereine) demonstrated significantly increased message levels for only 3 of 15 putative cytochrome P450s. Only one of the 3 senita alkaloid-inducible sequences, however, appeared to be a member of the CYP6A family, based on its strong sequence identity of 69% and 64% when aligned with D. melanogaster CYP6A9 and Ceratitis capitata CYP6A10, respectively. The corresponding region (helix L through the translational stop codon) of the remaining two senita alkaloid-responsive sequences displayed only weak sequence identity when aligned with known cytochrome P450 genes (e.g., 33% identity with CYP6B4v1).
Full-length cDNA sequences for these genes, CYP28A1 and CYP28A2, were subsequently obtained, and their conceptual amino acid translations reveal proteins clearly having the three salient characteristics of members of the cytochrome P450 superfamily. These are as follows: the conserved FXXGXXXCXG sequence of the heme-binding decapeptide near the C terminus, the DGXXT motif associated with formation of the oxygen-binding pocket of helix I (30), and the conserved EXXR pair within helix K that is thought to hydrogen bond with the “meander,” a stretch of residues N-terminal to the heme-binding decapeptide (31).
Historically, cytochrome P450 nomenclature has been based on a seemingly arbitrary set of criteria where proteins exhibiting <40% amino acid sequence identity have been assigned to separate gene families. In recent years, however, the cloning of cytochrome P450s from more divergent species, including arthropods, has necessitated a shift to a nomenclature system that focuses more on regional sequence identity and that takes into account the results of phylogenetic analyses. The cockroach CYP4C1, therefore, was included in the CYP4 family despite having only 32–36% positional identity with its vertebrate homologs (32). Similarly, conservation around the heme-binding decapeptide shows the evolutionary relatedness of CYP6A1 and CYP6B2 despite their <40% global sequence identity (2). The members of the CYP28 family, therefore, represent a distinct branch of xenobiotic-inducible cytochrome P450s. Evidence for the uniqueness of this gene family is found in the single most parsimonious tree of the aligned full-length sequences for 11 insect and 3 vertebrate cytochrome P450s, most of which have been linked to xenobiotic metabolism (Fig. 3). The clustering of the CYP28 sequence in the same clade as the xenobiotic-metabolizing vertebrate CYP3 and invertebrate CYP6 cytochrome P450s, rather than with the invertebrate CYP4 P450s [which are thought to play a role in energy mobilization rather than toxin metabolism (32)], suggest both evolutionary relatedness and the possibility of similar physiological functions. At the same time, bootstrap analyses point to the distinctiveness of the CYP28 sequences among xenobiotic-metabolizing families. When CYP4C1 was used as an outgroup, the CYP6 and CYP28 sequences in Fig. 3 formed separate monophyletic clades with bootstrap values of 80 and 100, respectively. Inclusion of the CYP4, CYP9, and CYP3 clades (each being monophyletic) with CYP11A1 as the outgroup produced a polytomy among all but the CYP4 sequences. This is consistent with the view that the major families of xenobiotic-metabolizing cytochrome P450s arose after the CYP4 family and suggests that the CYP28 family may be as distant from the invertebrate toxin-metabolizing enzymes as it is from vertebrate CYP3 cytochrome P450s.
The induction of the D. mettleri DU108 and CYP28 cytochrome P450s by the toxic alkaloids present in the tissue of one of the species’ host plants (i.e., senita cactus) and phenobarbital corroborate the results of earlier in vitro alkaloid metabolism studies (8, 17). These studies demonstrated a significant increase in alkaloid metabolism by isolated microsomes from D. mettleri larvae exposed to senita tissue and an even greater increase following phenobarbital exposure. Of greater significance with respect to possible involvement of these cytochrome P450s in xenobiotic metabolism, though, was the strong and highly specific induction of CYP28A1 by alkaloids that are present in senita cactus tissue at concentrations well in excess of those for any alkaloid in any other species of columnar cacti (i.e., 15–23% of the plant’s dry weight). Interestingly, exposure to an equivalent concentration of saguaro alkaloids did not produce a significant increase in CYP28A1 message. This is in sharp contrast to CYP28A2 and DU108, which were both crossinducible by senita as well as the chemically similar saguaro cactus alkaloids. This pattern suggests that resistance to senita alkaloids in D. mettleri may involve the cooperative action of multiple cytochrome P450s, which include at least one “specialist” that responds only to senita alkaloids and multiple “generalists” capable of responding to a broader range of chemically similar compounds. The action of such alkaloid-inducible “generalist” cytochrome P450s may also be reflected in the greater in vivo resistance of D. mettleri to another plant alkaloid, nicotine, relative to a non-alkaloid-resistant strain of D. melanogaster (33).
Finally, none of the 15 D. mettleri cytochrome P450s were induced by toxic triterpene glycosides isolated from the agria cactus. Insofar as induction has been indicative of enzymatic activity, the absence of induction suggests the lack of involvement of these genes in triterpene glycoside metabolism. With respect to the senita alkaloid-responsive cytochrome P450s, in particular, this was expected, given the chemical dissimilarity between isoquinoline alkaloids and triterpene glycosides. The general lack of triterpene glycoside inducibility was also not surprising, as D. mettleri larvae are unable to utlilize agria rot exudate-soaked soils.
CYP28A3 and CYP28A4 (isolated from D. nigrospiracula and D. hydei, respectively) represent two additional members of the CYP28 family. It appears more likely that both are orthologs of CYP28A1 rather than CYP28A2, in view of the greater positional identity of these genes vis-à-vis CYP28A1. Because D. nigrospiracula utilizes alkaloid-containing saguaro cactus tissue as a feeding and breeding substrate, it was possible to begin to determine whether the xenobiotic responsiveness of the CYP28 genes is a feature unique to D. mettleri or a more generalized characteristic of the gene family as a whole. A total of eight different D. nigrospiracula cytochrome P450 genes were screened by Northern hybridization for induction by saguaro alkaloids, triterpene glycosides, and phenobarbital. The general pattern of induction was remarkably similar to that of the CYP28 cytochrome P450s in D. mettleri. Phenobarbital induction of CYP28A3 was greater than for any other D. nigrospiracula cytochrome P450 tested. Furthermore, it was the only one of eight cytochrome P450s cloned from D. nigrospiracula to show induction by saguaro alkaloids. No induction by triterpene glycosides was observed. As before, these findings are consistent with previous in vitro alkaloid metabolism studies and the inability of D. nigrospiracula to utilize the triterpene glycoside-containing agria cactus as a host plant.
The demonstrated xenobiotic-inducibility of at least three members of the CYP28 family certainly points to a potentially significant role for the CYP28 family in host-plant utilization. At the same time, however, D. nigrospiracula clearly expresses an inducible CYP28 gene but is unable to utilize senita as a host plant, and D. hydei is incapable of utilizing any alkaloid-containing cactus species. Thus, while the CYP28 family, as a whole, may be readily induced by and possibly capable of conferring resistance to some plant allelochemicals, coevolutionary processes unique to the natural history of each species may account for the obvious differences in the observed patterns and magnitude of induction.
In conclusion, the use of a PCR-based cloning strategy has resulted in the isolation of four members of a previously unknown family of insect cytochrome P450s, designated CYP28, from three species of Drosophila. The CYP28 family is one of only two families of insect P450s (the other being CYP6) that have been found to display xenobiotic responsiveness at the mRNA level and may, therefore, play a role in the detoxification of natural and/or anthropogenic compounds. The ability of toxic isoquinoline alkaloids and phenobarbital to induce CYP28 message significantly in D. mettleri larvae suggests that (along with at least one other CYP6 cytochrome P450) these genes may be involved in utilization of soils soaked by senita cactus rot exudate. Similarly, at least one CYP28 gene may be a significant factor in saguaro cactus utilization by D. nigrospiracula.
Acknowledgments
This work was supported by National Science Foundation Grant DEB-9317885 and U.S. Department of Agriculture Grant 9304021 to J.C.F. and by National Science Foundation Grant DEB-9317884 to R.J.M.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RACE, rapid amplification of cDNA ends; rp49, ribosomal protein 49.
References
- 1.Omura T, Sato R. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 2.Nelson D R, Kamataki T, Waxman D J, Guengerich F P, Estabrook R W, Feyereisen R, Gonzalez F J, Coon M J, Gunsalus I C, Gotoh O, Okuda K, Nebert D W. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 3.Feyereisen R. In: Cytochrome P450. Schenkman J B, Greim H, editors. Heidelberg: Springer; 1993. pp. 311–324. [Google Scholar]
- 4.Grieneisen M L, Warren J T, Gilbert L I. Insect Biochem Mol Biol. 1993;24:115–132. doi: 10.1016/0965-1748(93)90077-6. [DOI] [PubMed] [Google Scholar]
- 5.Hodgson, E. & Rose, R. (1991) in Molecular Aspects of Monooxygenases and Bioactivation of Toxic Compounds, NATO Advanced Science Institutes Series, ed. Arinc, E. (Plenum, New York), pp. 75–91.
- 6.Tsukamoto M. In: Pest Resistance to Pesticides. Georghiou G P, Saito T, editors. New York: Plenum; 1983. pp. 71–98. [Google Scholar]
- 7.Oppenoorth F J. In: Comprehensive Insect Physiology, Biochemistry and Pharmacology. Kerkut G A, Gilbert L I, editors. Vol. 12. Oxford: Pergamon; 1985. pp. 731–744. [Google Scholar]
- 8.Frank M R, Fogleman J C. Proc Natl Acad Sci USA. 1992;89:11918–12002. [Google Scholar]
- 9.Berenbaum, M. R., Cohen, M. B. & Schuler, M. A. (1992) in Molecular Basis of Insecticide Resistance: Diversity among Insects, American Chemical Society Symposium Series 505, eds. Mullin, C. J. & Scott, J. G. (Am. Chem. Soc., Washington, DC), pp. 114–124.
- 10.Andersen J F, Utermohlen J G, Feyereisen R. Biochemistry. 1994;33:2171–2177. doi: 10.1021/bi00174a025. [DOI] [PubMed] [Google Scholar]
- 11.Tomita T, Scott J G. Insect Biochem Mol Biol. 1995;25:275–283. doi: 10.1016/0965-1748(94)00066-q. [DOI] [PubMed] [Google Scholar]
- 12.Saner C, Weibel B, Wuergler F E, Sengstag C. Environ Mol Mutagen. 1996;27:46–58. doi: 10.1002/(SICI)1098-2280(1996)27:1<46::AID-EM7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez F J, Nebert D W. Trends Genet. 1990;6:182–186. doi: 10.1016/0168-9525(90)90174-5. [DOI] [PubMed] [Google Scholar]
- 14.Ma R, Cohen M B, Berenbaum M R, Schuler M A. Arch Biochem Biophys. 1994;310:332–340. doi: 10.1006/abbi.1994.1175. [DOI] [PubMed] [Google Scholar]
- 15.Heed W B. In: Genetics, Speciation and the Founder Principle. Giddings L V, Kaneshiro K Y, Anderson W W, editors. New York: Oxford Univ. Press; 1989. pp. 253–278. [Google Scholar]
- 16.Kircher H W. In: Ecological Genetics and Evolution: The Cactus-Yeast Drosophila Model System. Barker J S F, Starmer W T, editors. Sydney, Australia: Academic; 1982. pp. 143–158. [Google Scholar]
- 17.Danielson P B, Frank M R, Fogleman J C. J Chem Ecol. 1994;20:1893–1906. doi: 10.1007/BF02066231. [DOI] [PubMed] [Google Scholar]
- 18.Meyer J M, Fogleman J C. J Chem Ecol. 1987;13:2069–2081. doi: 10.1007/BF01012872. [DOI] [PubMed] [Google Scholar]
- 19.Fogleman J C, Armstrong L. J Chem Ecol. 1989;15:663–676. doi: 10.1007/BF01014709. [DOI] [PubMed] [Google Scholar]
- 20.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skuse G R, Sullivan D T. EMBO J. 1985;4:2275–2280. doi: 10.1002/j.1460-2075.1985.tb03926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobsen K S, Haugen M, Sæbøe-Larsen S, Hollung K, Espelund M, Hornes E. In: Advances in Biomagnetic Separation. Uhlén M, Hornes E, Olsvik Ø, editors. Oslo: Eaton; 1994. pp. 61–71. [Google Scholar]
- 23.Schmidt T, Baker J T. Mech Ageing Dev. 1979;11:105–112. doi: 10.1016/0047-6374(79)90028-9. [DOI] [PubMed] [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Engler-Blum G, Meier M, Frank J, Müller G A. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 27.Juretic N, Theus M. FEBS J. 1991;290:4–8. doi: 10.1016/0014-5793(91)81212-q. [DOI] [PubMed] [Google Scholar]
- 28.Remmer H, Merker H J. Science. 1963;142:1657–1658. doi: 10.1126/science.142.3600.1657. [DOI] [PubMed] [Google Scholar]
- 29.Guengerich F P, Gillam E M J, Martin M V, Baba T, Kim B R, Shimada T, Raney K D, Yun C H. Schering Foundation Workshop, Assessment of the Use of Single Cytochrome P450 Enzymes in Drug Research. Berlin: Springer; 1994. pp. 161–186. [Google Scholar]
- 30.von Wachenfeldt C, Johnson E F. In: Cytochrome P450 Structure, Mechanism, and Biochemistry. 2nd Ed. Ortiz de Montellano P R, editor. New York: Plenum; 1995. pp. 183–223. [Google Scholar]
- 31.Peterson J A, Graham-Lorence S E. In: Cytochrome P450 Structure, Mechanism, and Biochemistry. 2nd Ed. Ortiz de Montellano P R, editor. New York: Plenum; 1995. pp. 151–180. [Google Scholar]
- 32.Bradfield J Y, Lee Y-H, Keeley L L. Proc Natl Acad Sci USA. 1991;88:4558–4562. doi: 10.1073/pnas.88.10.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danielson P B, Gloor S L, Roush R T, Fogleman J C. Pest Biochem Physiol. 1996;55:172–179. [Google Scholar]