Abstract
Platelet activating factor acetylhydrolase (paf-ah), a potent regulator of platelet activating factor activity, plays an important role in various physiological and pathophysiological functions including development, reproduction, inflammation, hemostasis, and apoptosis. Intracellular paf-ah (paf-ah-Ib) is composed of a regulatory subunit, Pafah1b1, and two highly conserved but non identical catalytic subunits, Pafah1b2 and Pafah1b3. The present study identifies new splice variants of the Pafah1b2 gene transcript. The splice variants retain exons 1–5 and replace exon 6 with alternative exons derived from genomic sequence 3′ to exon 6. Splice variants encode two proteins with different novel carboxy termini. One of the isoforms is expressed exclusively in testis. These new isoforms of pafah1b2 retain the ability to form higher order complexes while replacing known key catalytic residues, which raises the possibility that they may alter the subunit composition and catalytic function of paf-ah-Ib.
Introduction
Platelet activating factor (PAF) is a powerful regulator of cellular function, acting in an autocrine, paracrine, or endocrine like manner (1). Binding of PAF to surface receptors activates intracellular second messenger signaling cascades, including protein kinase C, tyrosine kinases, and mitogen-activated protein kinases (MAPKs), depending on target cell type (2, 3). PAF’s actions are not limited to receptor interactions at the cell surface, as it is internalized either bound to receptors or via an as yet uncharacterized non receptor mediated pathway (4, 5). Intracellular binding sites for PAF have been found in brain microsomal fractions (6) including a high affinity binding site implicated in transcriptional regulation (7). However, the role of PAF within the cytoplasm is still not well defined.
The PAF signal is modulated by platelet activating factor acetylhydrolases (paf-ah) that terminate the PAF signal by hydrolyzing the acyl group in the Sn2 position from the glycerol backbone (8). Three mammalian forms of paf-ah have been identified, a plasma isoform and two distinct intracellular enzymes designated types I and II (8, 9). The plasma and intracellular type II paf-ah isoforms have similar structures, 41% of amino acid residues are identical, and broad substrate specificities permitting them to act in an anti-inflammatory manner by inactivating a number of oxidized lipids in addition to PAF (8, 10).
In contrast to the plasma and intracellular type II isoforms, intracellular type I paf-ah (paf-ah-Ib) has a very different structure and a restricted substrate specificity that does not include oxidized lipids (8). This narrow substrate specificity is more consistent with regulatory enzymes in signaling pathways (11) than a generalized scavenger of reactive lipids. In addition to PAF, other structurally related lipids such as 1-OAlkyl-2-acetyl-sn-glycero-3-phosphoric acid (AAGPA-an intermediate in PAF synthesis) and 1-O-Alkyl-2-acetyl-sn-glycero-3-phosphatidylethanolamine (AAGPE) may act as second messengers whose signals are terminated by intracellular paf-ah-Ib catalyzed hydrolysis (12).
Paf-ah-Ib is a G protein-like trimer composed of two catalytic alpha subunits, Alpha-1 (Pafah1b3, 29 kDa) and Alpha-2 (Pafah1b2, 30kDa), and a beta regulatory subunit (Pafah1b1, LIS1, 45kDa) (11). The alpha subunits form a catalytic dimer, the activity of which is modulated by, but not dependant upon, association with the beta subunit. Alpha subunit composition of the dimer varies depending on tissue type and developmental stage (12, 13). Alpha-2 expression is ubiquitous, whereas Alpha-1 has a more tissue specific and developmentally regulated pattern of expression (13, 9).
The activity and substrate specificity of paf-ah-Ib is altered by the alpha subunit composition. The Alpha-2 homodimer and the Alpha-1/Alpha-2 heterodimer both have higher rates of PAF hydrolysis than the Alpha-1 homo-dimer (12). The regulatory effect of the beta subunit on paf-ah-Ib activity is also dependent on subunit composition. Association of the beta subunit with an Alpha-2 homodimer increases catalytic activity, whereas the interaction of the beta subunit with an Alpha-1 homodimer reduces catalytic activity (12, 9). The beta subunit has no effect on the activity of the Alpha-1 Alpha-2 heterodimer (12, 9).
The beta subunit (LIS1) also interacts, independently of the alpha subunits, with intracellular proteins including dynin, alpha tubulin, and nuclear distribution protein NUDE affecting a number of cellular processes involved in microtubule organization and transport (9). It has been suggested that the alpha subunits of paf-ah-Ib may also effect other intracellular processes utilizing properties unrelated to their catalytic capabilities (14).
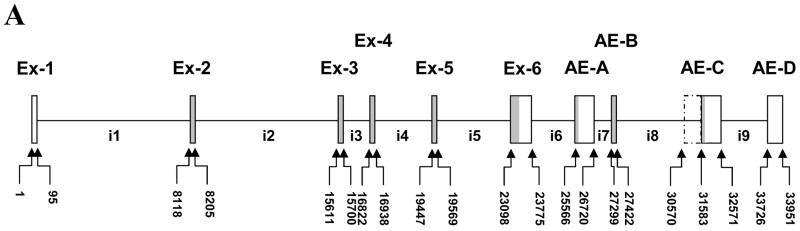
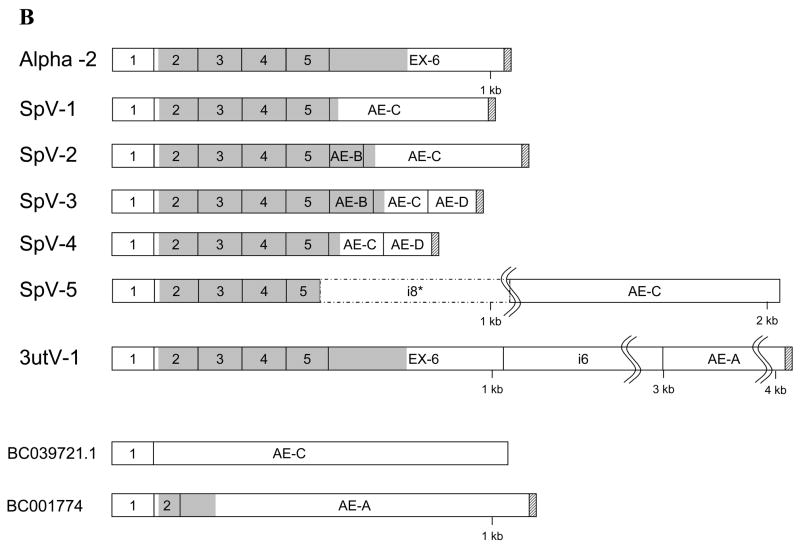
The sequence of human Pafah1b2 mRNA has been determined from cDNA cloning (13). The Genbank cDNA reference sequence (gi|4505584|ref|NM_002572.1) for the Pafah1b2 transcript is 1,195 nucleotides (not including the poly-A sequence). However, a transcript size of 4kb was reported in a tissue distribution study utilizing Northern blots (13). The corresponding human genomic sequence for Pafah1b2 consists of 6 exons (Figure 1) spanning approximately 23.77 kb (without 5′ and 3′ flanking regions) (15). Exons 2 through 6 encode the 229 amino acid catalytic Alpha-2 subunit of intracellular Paf-ah-Ib. Two alternatively spliced cDNA clones having identity to exon 1 of Pafah1b2 (BC039721.1) or exon1 and part of exon 2 (BC001774) spliced to differing 3′ sequences have been reported (16) (Figure 1 B). Both of these alternatively spliced mRNAs have open reading frames that would encode proteins having little (BC001774) or no identity (BC039721.1) with the canonical Alpha-2 subunit (Figure 1 panel B).
Figure 1.
Alternatively Spliced Pafah1b2 transcripts. Panel A, Exon organization of the Pafah1b2 gene. Panel B, schematic representations of the Pafah1b2 RNA splice variants. Canonical Alpha-2 exons are indicated by number with the prefix Ex-, introns are designated with “i” followed by a number, and alternate exons are indicated with the prefix AE followed by a letter. Identifying numbers beginning with BC are used for alternatively spliced clone sequences (16) obtained from GenBank. Coding regions are shaded, non genomic poly-A tails are indicated with diagonal striped fill. The portion of intron 8 included in SpV-5 is indicated with a dashed line. Exon sizes are representative, not drawn to exact scale. In panel B, introns and exons that were scaled grossly smaller to reduce figure size are marked with “S” shaped lines to note the scale change.
The current study describes novel Pafah1b2 RNA splice variants that maintain the canonical Alpha-2 configuration for exons 1 through 5 but replace exon 6 with alternate 3′ exons derived from the flanking region 3′ to exon 6. These new splice variants encode two protein isoforms that are identical to the catalytic Alpha-2 protein for their first 137 amino acids but have novel carboxy termini that replace several key catalytic residues.
Materials and Methods
cDNA Synthesis
First strand cDNA synthesis of human total RNAs for amplification using internal primers was done using 1ug total mRNA, 25nM Oligo dT18 (MWG), RNase inhibitor (Super RNase-in™ Ambion Cat # 2694), and Powerscript™ reverse transcriptase according to the enzyme manufacturer’s protocol (Clontech, Mountain View, CA). First strand cDNA synthesis for 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments were done using primers and protocol from SMART RACE amplification kit (Clontech, Mountain View, CA). In some cases the primer concentrations were reduced to 25nM for the Smart II A oligo and poly–A directed primers. All human total RNAs were obtained from Clontech (Mountain View, CA)
Amplification of Pafah1b2 cDNAs
Amplification of cDNA from exon 1 to the various alternate exons was accomplished using oligonucleotide primers selected from publicly available gene sequence (15, 17) and synthesized commercially (MWG-Biotech Inc. High Point, NC). Primer sequences and locations are given in Table 1. Amplifications were performed on first stranded cDNA templates made from human total RNA, as described above, or poly-A+ mRNA obtained from a commercial source (Clontech, Mountain View, CA). Advantage 2™ polymerase and the manufacturer’s recommended conditions (Clontech, Mountain View, CA Ref #639201) were employed for amplification. Amplification in a PTC-200 thermal cycler (MJ Research/BioRad Waltham, MA) required a touchdown thermal-cycling program (94°C 4.5 minutes, followed by 30 seconds at 94 °C \ 4 minutes at 72 °C for 5 cycles; 30 seconds at 94 °C \ 4 minutes at 70 °C for 5 cycles; 30 seconds at 94 °C \ 4 minutes at 68 °C for 25 cycles; 10 minutes at 68°C).
Table 1.
Sequences and locations for primers used in amplification and RACE experiments.
| Primer | Sequence | Location |
|---|---|---|
| 41-f | 5′ GGGACCGAGCGAGCGACCGAC 3′ | Ex-1 |
| 19515-f | 5′ CAGCAGAAGAAGTAGCAGGTGGGATCGAG 3′ | Ex-5 |
| 23671-r | 5′ CTTCATATTGTTCAGTATTTAGGCTTGATGTTGTCC 3′ | Ex-6 |
| 25188-f | 5′ GTTGGAGGAGGGCTTACTGATGCGTG 3′ | i6 |
| 26537-r | 5′ CAAGAGGCCCTAACTCACTTCTCCCACA 3′ | AE-A |
| 27330-f | 5′ AAAGCAGCCGCCTCCAAATACTCTATCTC 3′ | AE-B |
| 27384-r | 5′ GCCTGGTAGTTGCTGGTGTGTCATCAG 3′ | AE-B |
| 30770-f | 5′ TCTTTGGTCCATTCAACACTCAGGCTCAG 3′ | i8 |
| 31723-r | 5′ CAGCACATCTAGGCGAATATGAATCAGAGAATC 3′ | AE-C |
| 31755-f | 5′ GATTCTCTGATTCATATTCGCCTAGATGTGCTG 3′ | AE-C |
| 31992-r | 5′ CTGTTGTAGTTATGCTGGGCAAATGGTGAG 3′ | AE-C |
| 33747-r | 5′ CAAGAAACTTAGAGAGCTTTATACCCTTTGATCCAGTA 3′ | AE-D |
| Long-UP | 5′ CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT 3′ | C |
| Short-UP | 5′ CTAATACGACTCACTATAGGGC 3′ | C |
| Nest-UP-A | 5′ AAGCAGTGGTATCAACGCAGAGT 3′ | C |
| SMART II A | 5′ AAGCAGTGGTATCAACGCAGAGTACGCGGG 3′ | C |
| 3′-RACE CDS | 5′ AAGCAGTGGTATCAACGCAGAGTAC(T)30VN 3′ | C |
| 5′-RACE CDS | 5′ TTTTTTTTTTTTTTTTTTTTTTTTTVN 3′ | C |
| T7-to-Met1-f | 5′ GGATCCTAATACGACTCACTATAGGGAGCCACCATGAGCCAAGGAG ACTCAAACCCAGCAGC 3′ | Ex-2 |
Key: Ex-1 = exon 1, Ex-5 = exon 5, Ex-6 = exon 6, AE-A = alternate exon A, AE-B = alternate exon B, AE-C = alternate exon C, i6 = intron 6, i8 = intron 8, and C = Clontech Smart RACE™ primer. Ambiguous nucleotide code is N = G, A, T, or C and V = G, A, or C.
5′ and 3′RACE
The 3′ and 5′ ends of splice variants first identified by amplification between exon specific primers were determined using RACE. Preliminary experiments using 3′ RACE primed in exon 5 produced multiple products. To enrich reactions in amplification products containing the target exon of interest, nested RACE reactions were designed in which the first amplification was primed in an exon flanking the exon of interest. The first amplification product was diluted and used as template for nested RACE reactions using a gene specific primer directed to the new exon. For 3′ RACE the primary amplification was usually initiated in exon 5. For some 5′ RACE reactions both primary and nesting primers were chosen within the terminal exon. The combined RACE experiments confirmed the sequence from the direct amplification using gene specific primers and added the 5′ and 3′ terminal sequences. Gene specific primer sequences are listed in Table 1 and primer combinations in Table 2. In some cases gel purified products were re-amplified to obtain larger quantities for sequencing.
Table 2.
Primer sets used for cDNA amplification (Part A), RACE experiments (Part B), and In Vitro translation (Part C).
| A. | |||||||
|---|---|---|---|---|---|---|---|
| Set | Target | Use | Forward | Reverse | RNA source | ||
| 1 | Ex-1 to AE-C | Amplification | 41-f | 31992-r | Tissues, 1st Strand cDNA | ||
| 2 | Ex-1 to AE-B | Amplification | 41-f | 27384-r | Tissues, 1st Strand cDNA | ||
| 3 | Ex-1 to AE-D | Amplification | 41-f | 33747-r | Tissues, 1st Strand cDNA | ||
| 4 | Ex-1 to AE-A | Amplification | 41-f | 26537-r | Tissues, 1st Strand cDNA | ||
| 5 | Ex-1 to Ex-6 | Amplification | 41-f | 23671-r | Tissues, 1st Strand cDNA | ||
| B. | |||||||
| RACE | RACE | Primary RACE Primers | Nesting RACE Primers | RNA | |||
| Set | Target | Type | Forward | Reverse | Forward | Reverse | source |
| 6 | AE-C/AE-D | 3′ RACE | 19515-f | UPM | 31755-f | NUP-A | Kidney/Testis |
| 7 | AE-C | 5′ RACE | UPM | 31992-r | NUP-A | 31723-r | Testis |
| 8 | AE-B | 3′ RACE | 19515-f | UPM | 27330-f | NUP-A | Testis |
| 9 | AE-B | 5′ RACE | UPM | 31723-r | NUP-A | 27384-r | Testis |
| 10 | AE-D | 3′ RACE | 19515-f | UPM | 27330-f | NUP-A | Testis |
| 11 | I8 (SpV-5) | 3′ RACE | 19515-f | UPM | 30770-f | NUP-A | Testis |
| 12 | AE-A | 3′ RACE | 19515-f | UPM | 25188-f | NUP-A | Testis |
| C. | |||||||
| Set | Target | Use | Forward | Reverse | Template | ||
| 13 | Ex-2 to Ex-6 | In Vitro Translation | T7-to-Met1-f | 23671-r | Purified Human Testis Alpha-2 cDNA | ||
| 14 | Ex-2 to AE-C | In Vitro Translation | T7-to-Met1-f | 31723-r | Purified Human Testis SpV-1 or SpV-2 cDNA | ||
Key: UPM = Universal primer mix; a mix of Long UP (0.4uM) and short UP (2uM) primers. NUP-A = Nest-UP-A
Amplifications for 5′ and 3′ RACE experiments were done using a kit (SMART RACE Clontech, Mountain View, CA) and first stranded cDNA made from human testis mRNA as described above. Primers for RACE experiments including kit primer sequences are given in Table 1. The manufacturer’s protocol was modified by decreasing the annealing temperatures of the touchdown amplification protocol (94°C 4.5 minutes, followed by 30 seconds at 94 °C \ 4 minutes at 72 °C for 4 cycles; 30 seconds at 94 °C \ 30 seconds at 65 °C \ 4 minutes at 68 °C, for 4 cycles; 30 seconds at 94 °C \ 30 seconds at 60 °C \ 4 minutes at 68 °C for 22 cycles; 10 minutes at 68°C).
DNA purification
Amplified products were separated by electrophoresis in agarose Tris/Borate/EDTA (TBE) gels and stained with ethidium bromide. Amplified product bands were excised from the gel and then purified using GeneClean™ turbo (Qbiogene, Irvine, CA) or Qiaquick™ (Qiagen Valencia, CA) spin columns. Purified products were quantified using a NanoDrop™ spectrophotometer (NanoDrop Technologies, Wilmington, DE).
DNA Sequencing
Sequencing reactions were performed using purified amplification products with Big Dye Terminator cycle sequencing reagents (Applied Biosystems, Foster City, CA). Completed sequencing reactions were purified using Centrisep 100 spin columns (Princeton Separations, Adelphia, NJ) according to the manufacturer’s protocol. Sequencing reactions were resolved on an ABI 3130xI sequencer (Applied Biosystems, Foster City, CA).
Sequence analysis
DNA sequence alignments and electropherogram viewing were done using Sequencher software (Gene Codes Corporation, Ann Arbor, MI). Protein secondary structure predictions were performed using Lasergene 6 sequence analysis software (DNASTAR, Madison, WI). DNA alignments, DNA homology searches, protein homology, and protein conserved domain searches were performed on line at the NCBI web site using Blast and CD-Search (http://www.ncbi.nlm.nih.gov/) (18). Phosphorylation site predictions were performed online using the online Eukaryotic Linear Motif Resource for functional sites in proteins (ELM) (http://www.elm.eu.org/) (19) in conjunction with NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos/) (20).
Tissue Distribution
To address the expression of alternatively spliced variants in various tissues, normalized cDNA panels from Clontech (MTC panel 1, K1420-1 Ref 636742 and panel 2, K1421-1) were used with Pafah1b2 primers that amplify between exon 1 and the alternative exons (Table 2). Amplification between exon 1 and exon 6 of Pafah1b2 was performed to show relative Alpha-2 tissue levels. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene expression was also determined on all samples as a normalization reference (G3PDH primers from Clontech, Mountain View, CA Cat #S1222). Pafah1b2 amplifications used the same touchdown program as given in the amplification of splice variants section above, with the only modification being that for amplifications between exon 1 and 6 the cycle number in the last segment was reduced from 25 to 21, to keep the amplification in log phase. The G3PDH amplification protocol was: one cycle of 95°C 4.5 minutes; 21 cycles of 30 seconds at 95 °C \ 3 minutes at 68 °C; and one cycle of 5 minutes at 68°C. All amplification reactions used 5ug of cDNA and the manufacturer’s recommended conditions for Advantage 2™ polymerase. To visualize the results, an equal volume of completed amplification reaction for each tissue/primer set was separated by electrophoresis in 1.5% agarose TBE gels and stained with ethidium bromide.
In Vitro Translations
Coupled in vitro transcription/translation reactions utilized a rabbit reticulocyte system (TnT™ T7 Quick for PCR DNA system for PCR, cat # L5540, Promega, Madison, WI) and amplified cDNA templates that inserted a T7 RNA polymerase promoter at the 5′end of the sense strand. The manufacturer’s protocols were followed for coupled in vitro transcription/translation reactions.
Templates for the transcription/translation reactions were constructed using polymerase chain reaction and a forward primer (T7-to-Met1-f, Table 1) that adds a T7 RNA polymerase promoter seven nucleotides before the initiating methionine of Pafah1b2. T7 promoter linked template for canonical Alpha-2 in vitro translation (IVT) was generated from primer set 13 (Table 2c) using the gel purified 4.1kb amplification product from primer set 4 as template (Table 2a). T7 promoter IVT templates for SpV-1 and SpV-2 were made in separate reactions using primer set 14 (Table 2c) with gel purified amplification products from primer set 1 (Table 2a) as templates, the 932 nucleotide product was used for SpV-1 and the 1,056 nucleotide product for SpV-2. The single strand cDNA used for all primary amplifications was made from human testis mRNA (Clontech, Mountain View, CA) as described above. T7 promoter addition and identity of purified IVT template cDNAs were confirmed by sequencing.
Western Blotting
Polyacrylamide gel electrophoresis (PAGE) of in vitro translated products and transfer to PVDF membranes was accomplished using the Novex™ pre-cast gel system from Invitrogen (Carlsbad,CA). Native, denaturing (SDS), and two dimensional Tris/Glycine gels were run and transferred to PVDF membranes according to the manufacturer’s protocols. Testis tissue extract (10μg/lane) was used as a reference sample (USBiological, Cat# T5595–6599, Swampscott, MA). The Pafah1b2 primary antibody was a chicken polyclonal obtained from Abcam (Cat # Ab 15875, Cambridge, MA). The secondary antibody was rabbit anti-chicken linked either to Horseradish Peroxidase (Cat # SJ31804) or Alkaline Phosphatase (Cat # SJ31805, Biomeda, Foster City, CA). Western Blue™ chromogenic substrate, and Transcend™ chemiluminescent substrate were obtained from Promega (Madison, WI). Immunostaining with chromogenic or chemiluminescent substrates was performed in accordance with the manufacturer’s protocols (Promega, Madison, WI). Kodak BioMax Light™ film (Cat#178 8207, Rochester, NY) was used for chemiluminescent detection. Sizes of protein products separated by PAGE were estimated using Quantity One™ software (Bio-Rad Laboratories, Hercules, CA).
Results
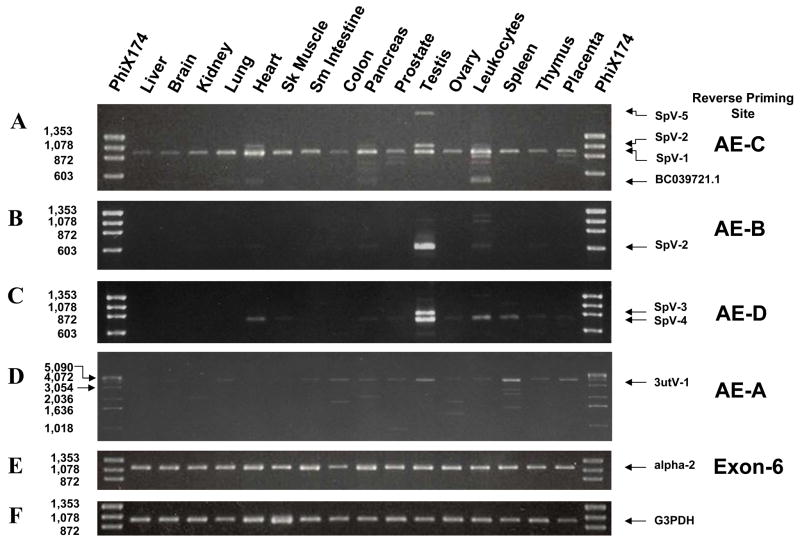
Novel alternatively spliced RNAs from the Pafah1b2 gene were identified by amplification and sequencing of first stranded cDNA using primers located in exon 1 and sequences located 3′ to exon 6. The sequence used for the 3′ primer was obtained from GenBank clone BC039721.1 that has exon 1 of pafah1b2 gene spliced to a distal 3′ flanking sequence (Figure 1, Table 2 primer set 1). The 3′ sequence of clone BC039721.1 is referred to herein as alternate exon C (AE-C). The number of amplification products varied with tissue type and ranged from a single product to more than six distinct bands (Figure 2 panel A). Three major amplification products of 932; 1,056; and 1,944 nucleotides represent novel splice variants, designated here as SpV-1, SpV-2 and SpV-5 respectively, are described below. Alternatively spliced sequences were confirmed and mRNA ends determined in RACE experiments for each splice variant. The RACE experiments also identified another exon (AED) found 3′ to AE-C that is carried by two additional splice variants, SpV-3 and SpV-4 (Figure 1). A 3′ untranslated sequence variant (3utV-1) is also described. There are additional low abundance amplification products that remain uncharacterized at this time. Sequences of characterized novel splice variants described below have been deposited in Genbank (accession numbers are; SpV-1 = DQ836742, SpV-2 = DQ836741, SpV-3 = DQ836740, SpV-4 = DQ836739, SpV-5 = DQ836738, 3utV-1 = DQ836743).
Figure 2.
Tissue distribution of Pafah1b2 splice variants. All amplifications utilized the same exon 1 forward primer paired with a reverse primer selected from the target exon. Panel A, Amplification between exon 1 and AE-C. Amplification products that have been characterized are indicated with arrows. Panel B, Exon 1 to AE-B amplification products. Panel C, Exon 1 to AE-D amplification products. Panel D Exon 1 to AE-A amplification products. Panel E, products from exon 1 to exon 6 amplification that represents the canonical Alpha-2 splice pattern. Panel F, Control amplification of the G3PDH transcript. Amplification conditions were adjusted to keep all reactions in log phase.
Description of the Alpha-2 cDNA Variants
Splice variant 1 (SpV-1), the 932 nucleotide product from primer set 1 (Table 2), was sequenced and found to link exons 1 through 5 to AE-C (Figure 1, Figure 2 panel A). The 3′ splice junction for exon 5 in SpV-1 is the same as used for exon 5 to exon 6 splice in the canonical Alpha-2 transcript. The 5′ splice junction of AE-C was the same as used for linking exon 1 to AE-C in clone BC039721.1 (Figure 1).
The original report of clone BC039721.1 included a 3′ terminal poly-A sequence that is of genomic origin and is associated with an Alu repeat sequence (Figure 3). Using 3′ RACE experiments initiated within exon 5 we were able to demonstrate that post transcriptional processing of AE-C in SpV-1 (Table 2 primer set 6) results in polyadenylation 571 nucleotides downstream from the stop codon following a type II (AUUAAA) signal (21) (Figure 3A). In SpV-1 the poly-A sequence is added before the aforementioned Alu repeat, 465 nucleotides upstream from the genomic poly-A tract of BC039721.1 (Figure 3). 5′ RACE experiments amplified from AE-C (Table 2 primer set 7) confirmed exon 1 to be the 5′terminus for SpV-1 mRNA. The transcript size for SpV-1 is 1,017 nucleotides including the 5′ and 3′ untranslated sequences but not the non-genomic poly-A sequence.
Figure 3.
Alignment of splice variant 3′ cDNA ends to Pafah1b2 genomic sequence showing poly-A addition sites for alternate exons AE-C, AE-A and AE-D as determined by 3′ RACE. Genomic sequence is shown in bold and polyadenylation signals are underlined. Panel A, SpV-1 and SpV-2 cDNA 3′ sequence (AE-C). Panel B, Genomic derived poly-A sequences found in SpV-5 and clone BC039721.1 aligned with genomic sequence. Panel C, Alignment of the 3′ end of 3utV-1 cDNA sequence (which is identical to the 3′ end of clone BC001774) (AE-A) with genomic sequence. Panel D, Alignment of the 3′ end of SpV-3 (AE-D) cDNA sequence with genomic sequence. Panel E, Alternate poly-A site seen in SpV-3 and SpV-4 (AE-D) aligned with genomic sequence.
Splice variant 2 (SpV-2) was discovered in testis cDNA as a 1,056 nucleotide product (Table 2 primer set 1, Figure 2 panel A). In SpV-2 exons 1 through 5 are joined with a novel exon, alternate exon B (AE-B), followed by alternate exon AE-C. AE-B is 124 nucleotides long and is located 4,153 nucleotides downstream from exon 6 in the pafa1b2 gene (Figure 1). The splice junctions for exons flanking AE-B, the 3′ site of exon 5 and 5′ site of AE-C, are the same as described for SpV-1. Splice sites of AE-B conform to normal junction architecture. The reading frame from exon 5 codes through AE-B to AE-C. The reading frame for AE-C seen in SpV-1, however, is not preserved in SpV-2, which results in different predictions for the carboxy terminal protein sequence from the same exon. The 5′ and 3′ ends and polyadenylation site of SpV-2 cDNA were confirmed by sequencing 5′ and 3′ RACE products amplified from within AE-B (Table 2 primer sets 8 and 9). SpV-2 uses the same polyadenylation site in AE-C as found for SpV-1 (Figure 3A). Transcript size including 5′ and 3′untranslated regions for SpV-2 is 1,141 nucleotides.
SpV-1 and SpV-2 encode proteins with different carboxy termini each unique to that of canonical Alpha-2. The additional splice variants that are described below alter the 3′ exon composition of the Pafah1b2 RNA but do not encode any protein variations in addition to those of SpV-1 or SpV-2.
Splice variant 3 (SpV-3) was identified in 3′ RACE experiments and splices exons 1 through 5 to AE-B, AE-C, and another new exon, AE-D. AE-D is approximately 240 nucleotides and is located in the genomic sequence 2,143 nucleotides downstream from the 3′ splice site of AE-C (Figure 1). In SpV-3, AE-D is spliced to AE-C 192 nucleotides downstream from the 5′ splice junction before the poly-A addition site described for SpV-1. The predicted protein for SpV-3 is the same as SpV-2 as AE-D only adds additional 3′ untranslated sequence.
Splice variant 4 (SpV-4) includes exons 1 through 5, AE-C, and AE-D and has the same splicing as SpV-1 with the 3′ addition of AE-D. The AE-C to AE-D splice junction is the same as seen in SpV-3. Amplification products from primer sets 3, 6, 8, and 10 (Table 2) were used to determine the sequences of SpV-3 and SpV-4. The protein encoded by SpV-4 is identical to that encoded by SpV-1.
A fraction of SpV-3 and SpV-4 transcripts show use of an alternate 3′ splice site in AE-C that is 23 nucleotides downstream (3′) from the splice junction described above. The two 3′ AE-C splice sites occur 119 and 142 nucleotides, respectively, downstream from the stop codon in AE-C. Sequencing of 3′ RACE products for SpV-3 and SpV-4 (Table 2 primer sets 8 and 10) was complicated by the use of these alternative 3′ splice junctions and by polyadenylation that occurs at two different sites separated by 21 nucleotides in AE-D (Figure 3). RACE experiments were, however, able to confirm non-genomic polyadenylation. No consensus polyadenylation signal sequences were found adjacent to the polyadenylation sites in AE-D. The approximate transcript size for SpV-3 is 950 nucleotides and 826 nucleotides for SpV-4; this includes the 5′ and 3′ untranslated regions but not the non-genomic poly-A tail.
Splice variant 5 (SpV-5), the 1,944 nucleotide amplification product seen in testis (Primer set 1 Table 2 and Figure 2 panel A), includes exons 1 through 4, most of exon 5, and AE-C. This variant utilizes a 5′ splice junction for AE-C located in intron 8 (i8) 1,009 nucleotides upstream from that used in all the other AE-C carrying splice variants. The 3′ splice site of exon 5 is 16 nucleotides upstream from its usual position (Figure 1). Use of these altered splice sites introduces a stop codon at the exon 5/i8-AE-C splice junction. This splice variant does not add any new amino acids, rather, the alternate splice results in a truncated protein ending at Ala132. RACE experiments (Table 2 primer set 11) determined SpV-5 ends in the same genomic poly-A sequence reported for clone BC039721.1 (Figure 3B), indicating that this may be an incompletely processed transcript. Predicted transcript size for SpV-5 is 2,474 nucleotides without the genomic poly-A sequence.
The sequence of clone BC001774 (16) has a splicing pattern of exon 1 spliced to a truncated exon 2 which is spliced to a 3′ flanking sequence occurring between exon 6 and AE-B. This 3′ flanking sequence is designated here as alternate exon A (AE-A) (Figure 1A and B). Amplifications between exon 1 and AE-A were performed in order to determine if alternate exon A (AE-A) was utilized in a larger more complete Alpha-2 transcript than seen in clone BC001774. Amplification with primer set 4 (Table 2) produced multiple low abundance tissue specific products of varying size (Figure 2 panel D) and a relatively abundant more widely expressed 4,111 nucleotide product (expected BC001774 product size was 1,090 nucleotides). Sequencing of the 4.1 kb cDNA and related 3′RACE products (Table 2 primer sets 4 and 12) showed it to have canonical Alpha-2 splicing linking exons 1 through 6 with the 3′ untranslated sequence extending from exon 6 to a non-genomic poly-A tail 2,949 nucleotides downstream from the polyadenylation site reported for the human Alpha-2 cDNA (13) (Figure 3). This 4.1kb cDNA, referred to herein as 3′ untranslated variant 1 (3utV-1) (Figure 1B), uses the same polyadenylation site as reported for clone BC001774 (Figure 3C). The 3utV-1 and the canonical Alpha-2 transcript (13) do not have common consensus polyadenylation signals but have type IV and type III elements, respectively (21). Predicted transcript size for the 3utV-1 is 4,136 nucleotides including the 5′ and 3′ untranslated regions. The 3utV-1 sequence encodes a canonical Alpha-2 subunit and may correlate to the 4 kb RNA reported for the Alpha-2 subunit in Northern Blots (13).
Tissue Distribution of Alpha-2 cDNA variants
The tissue distribution of splice variants was determined using a qualitative amplification of first strand cDNA with a forward primer located in exon 1 and a reverse primer in the alternate exon of interest (Table 2 primer sets 1–5). The number of products and pattern of expression are quite different for each of the alternate exons.
SpV-1 is ubiquitously expressed (Figure 2 panel A) and in many tissues is the only product that contains AE-C. The level of SpV-1 expression, however, varies between the tissues and does not mirror the relatively uniform expression of the catalytic Alpha-2 transcript (Figure 2 panel E).
Products representing SpV-2 and SpV-3 are predominantly expressed in testis with only trace amounts of SpV-2 seen in leukocytes, heart, and pancreas (Figure 2 panels A, B, and C). SpV-4 has its highest expression in testis but also is expressed in heart, leukocyte, and spleen (Figure 2 panel C). Trace levels of SpV-4 expression was also seen in several other tissues. SpV-5 was seen only in testis (Figure 2 panel A).
Expression of the 3′ untranslated variant 3utV-1, seen as a 4.1kb product, was found in most tissues with the highest levels in spleen and testis. Multiple minor products were also seen in the exon 1 to AE-A amplification that are expressed in a tissue specific manner, but have not been characterized further.
Predicted Protein Sequences of Alpha-2 Splice Variants SpV-1 and SpV-2
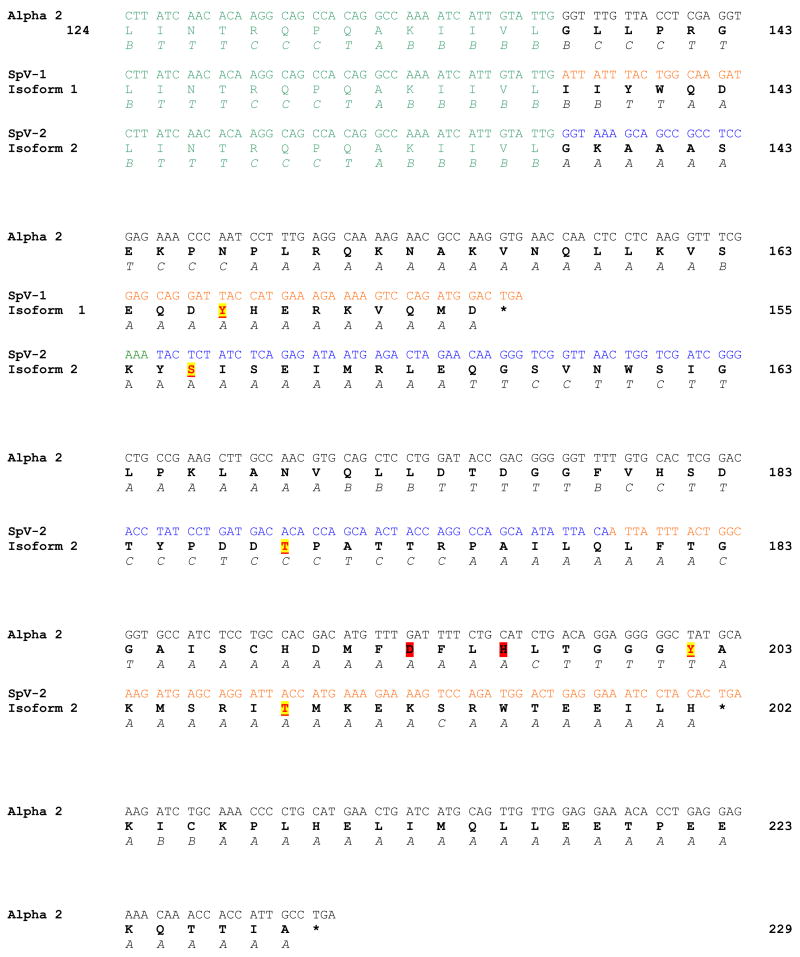
The predicted protein sequence from SpV-1 cDNA is 155 amino acids, with a molecular mass of ~17.8 kDa. Amino acids 1–137 are identical to the original 229 amino acid canonical form of Pafah1b2 (Alpha-2), and the 95 amino acids encoded by exon 6 in Alpha-2 are replaced. The carboxy terminus of SpV-1 is composed of 18 amino acids that are encoded by AE-C with the reading frame defined by exon 5 (Figure 4). The predicted structure of the alternate carboxy terminus in SpV-1 is primarily hydrophilic alpha helix with a high surface probability.
Figure 4.
Carboxy terminal sequences with protein translations, secondary structure predictions, and phosphorylation sites for Alpha-2, SpV-1 (protein isoform 1) and SpV-2 (protein isoform 2). Garnier-Robbins secondary structure predications Alpha helix (A), Beta Sheet (B), Coil (C), and Turn (T) are shown below the protein sequence. Predicted phosphorylation sites are underlined and highlighted in yellow. Catalytic triad residues Asp193 and His196 in Alpha-2 are highlighted in red. Common sequence encoded by exon 5 is in green, exon 6 cDNA sequence (Alpha-2) is in black, AE-C sequence is in orange, and AE-B cDNA sequence is in blue. Amino acids 1–137 are identical for all three proteins; residues 1–123 are not shown. Stop codons are marked with *.
In the SpV-2 transcript canonical residues 1–137 are followed by a unique sequence of 65 amino acids resulting in a predicted protein sequence that is 202 amino acids with a molecular mass of ~22.8 kDa. The new carboxy terminal amino acid sequence is predicted to begin with an amphipathic alpha helix, followed by a central region composed of coil and turn, and ending with a hydrophilic alpha helix (Figure 4).
The novel carboxy terminal sequences of SpV-1 or SpV-2 did not show homology with other proteins in Genbank. A conserved domain structure search also failed to find similarities with known protein domains. There were, however, predictions for phosphorylation within the novel carboxy termini encoded by alternate exons AE-B and AE-C. The 18 amino acid carboxy sequence encoded by SpV-1, has a potential Tyrosine phosphorylation site at Tyr147 for epidermal growth factor receptor (EGFR) (Figure 4) (19). SpV-2 carboxy terminal sequence has phosphorylation sites predicted for casein kinase I (CKI) and casein kinase II (CKII), at Ser146, mitogen activated protein kinase (MAPK) at Thr169, and protein kinase C (PKC) at Thr189 (SpV-2 numbering).
Expression of Alpha-2 Isoforms SpV-1 and SpV-2 In-Vitro
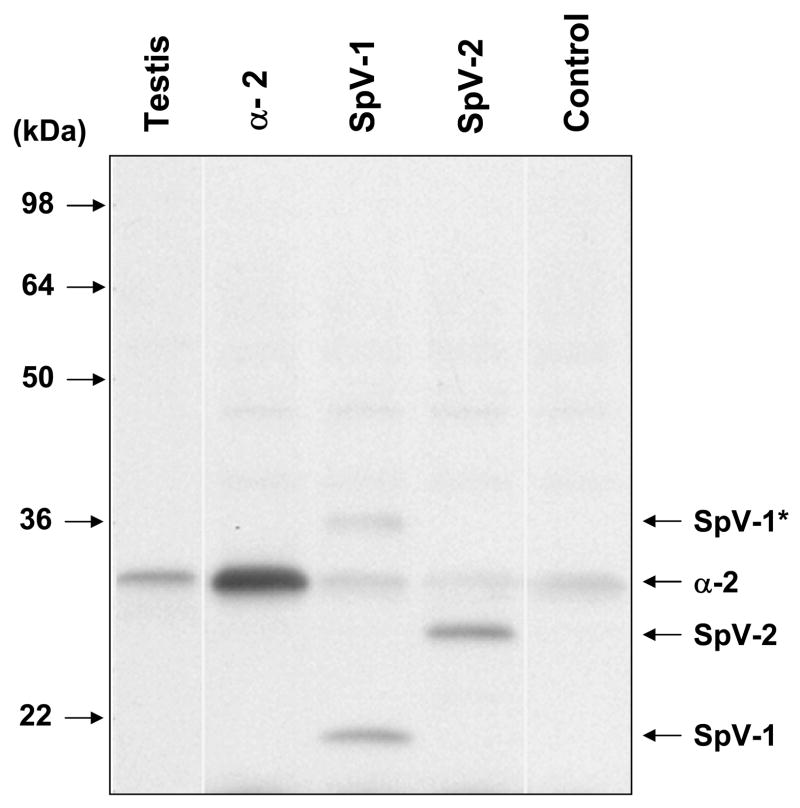
In-vitro translation of canonical alpha subunits (Alpha-1 and Alpha-2) yielded protein products in the expected size range. Paf-ah-Ib alpha subunits are known to run anomalously in SDS-PAGE, for example, the Alpha-2 subunit has a predicted molecular weight of 25.5 kDa but runs at ~30kDa in SDS-PAGE (22). The mobility of the splice variants in SDS-PAGE were also larger than predicted, with in-vitro translated SpV-2 running at ~25kDa instead of 22.8kDa (Figure 5). In-vitro translated SpV-1, however, was seen as a ~35kDa band, about double the predicted 17.8kDa expected, suggesting that SpV-1 may form a relatively stable dimer. Under strong denaturing conditions the ~35kDa band is disrupted and a ~19kDa band is seen that is closer to the predicted SpV-1 size (Figure 5). Denaturing conditions in Figure 5 were chosen to partially disassociate the 35kDa band so both forms of SpV-1 could be shown. A less abundant SpV-1 complex was seen at ~62kDa (data not shown). No bands were seen in the rabbit reticulocyte lysate control translation lane (Lane 5, Control) of a size similar to SpV-1 or SpV-2; however, endogenous rabbit Alpha-2 is detected. Although the polyclonal Pafah1b2 antibody was raised to canonical Alpha-2 it also recognizes SpV-1 and SpV2 suggesting that there are epitopes bound by the antibody that lie in the first 137 amino acids, which are identical for all three of these proteins.
Figure 5.
Pafah1b2 immunostain of in-vitro translated Alpha-2, SpV-1, and SpV-2 separated in a denaturing Tris/Glycine 10–20% acrylamide SDS-PAGE system. In-vitro translations were performed in rabbit reticulocyte lysates using purified cDNA templates produced from human testis RNA. The completed translations were loaded directly on the gel without purifying protein products from the lysate. Chemiluminescent immunostaining is with Pafah1b2 polyclonal primary antibody and a horseradish peroxidase linked secondary. Lane 2 (α2) shows translation from human canonical Pafah1b2 template. Note: endogenous rabbit Pafah1b2 is seen in the control and splice variant translations. SpV-1 and SpV-2 monomers are indicated with labeled arrows. The band corresponding to non-disassociated SpV-1 complex is labeled as SpV-1*. Testis tissue sample (Lane 1) is a total cell lysate of human origin. Control sample is a rabbit reticulocyte lysate transcription/translation reaction performed in the absence of exogenous template.
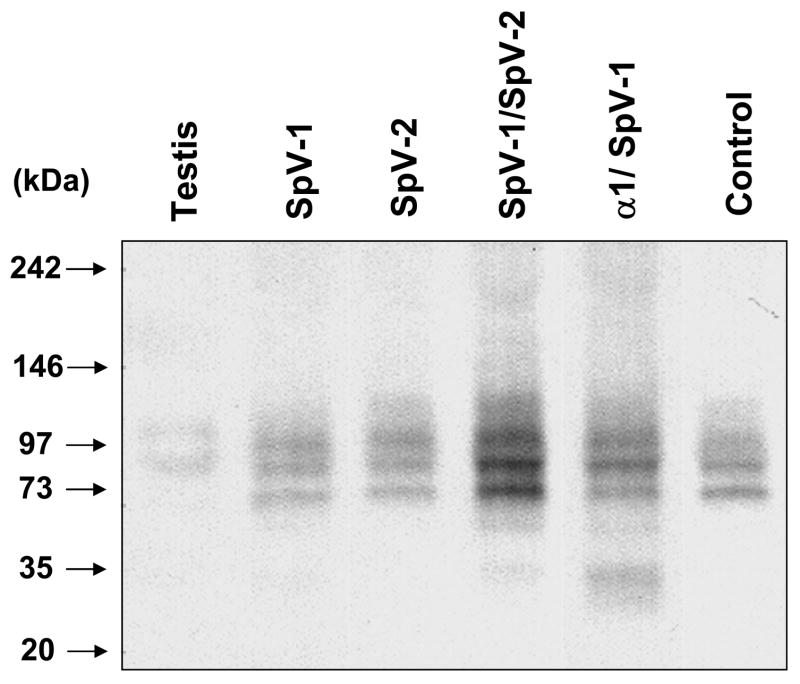
Native gel electrophoresis of in-vitro translated SpV-1 shows that it forms other higher molecular weight complexes ranging from 65kDa to 110kDa in addition to the ~35kDa complex (Figure 6). In-vitro translated SpV-2 also appears as higher molecular weight complexes in native gel electrophoresis, with a band at 120kDa and enrichment of an endogenous Pafah1b2 band at 98kDa. The predicted molecular weight of a SpV-2 homo-dimer is ~45kDa and an Alpha-2/SpV-2 hetero-dimer is ~48kDa. No signal in the monomeric size range for SpV-1 or SpV-2 was seen in native gels (Figure 6). Figure 6 also shows that endogenous Pafah1b2 in the rabbit reticulocyte lysate (Control lane) runs as two relatively abundant complexes at 74kDa, and 86 kDa, a less abundant band at 98 kDa, and a diffuse band of about 115kDa. The human testis tissue extract sample (Figure 6, Testis lane) had two prominent bands, one at ~85kDa and another at 105kDa. The predicted sizes of canonical Paf-ah-Ib complexes, using their combined molecular weights, are ~51 kDa for alpha subunits in a homo or hetero-dimer configuration and ~98kDa for the Alpha-1, Alpha-2, and beta subunit hetero-trimer. There were no prominent bands seen in the expected size range for alpha subunit monomers or dimers in either the testis tissue sample or the reticulocyte lysate control translation. Both the rabbit reticulocyte lysate and the human testis tissue sample had additional bands that are of unknown composition but have components that are bound by the pafah1b2 specific antibody. Some of the reticulocyte lysate endogenous bands are enhanced and their mobility slightly altered (+ 1 to 5 kDa depending on the band) by translation of the splice variants. It may be that this enhancement of endogenous bands is due to the exchange of in-vitro translated SpV-1 and/or SpV-2 for endogenous alpha subunits within these complexes.
Figure 6.
Native gel electrophoresis of unpurified in-vitro translated products separated in a non-denaturing Tris/Glycine 4–12% acrylamide Native-PAGE system. Chromogenic immunostaining is with Pafah1b2 polyclonal primary antibody and an alkaline phosphatase linked secondary. Testis sample is of human origin. Control sample is a transcription/translation reaction performed in the absence of exogenous template.
Co-translation of SpV-1 with SpV-2 or Alpha-1 increased yields and resulted in the formation of complexes consistent with hetero-dimers and higher order complexes (Figure 6). SpV-1 co-translated with Alpha-1 gave a rather prominent doublet at 29kDa and 35kDa, faint bands at ~55kDa and 118kDa, and enhancement of an 84kDa band. Whereas co-translation of SpV-1 with SpV-2 gave bands at 38kDa, 63kDa and 120 kDa, most of the product was seen as enhancement of 74kDa and 86kDa bands. The predicted molecular weight of a SpV-1/Alpha-1 hetero-dimer is ~43kDa and for a SpV-1/SpV-2 hetero-dimer is ~40kDa. The Pafah1b2 antibody does not recognize Alpha-1. Co-translation of SpV-1 or SpV-2 with canonical Alpha-2 did not produce banding dramatically different than the combined individual patterns (data not shown).
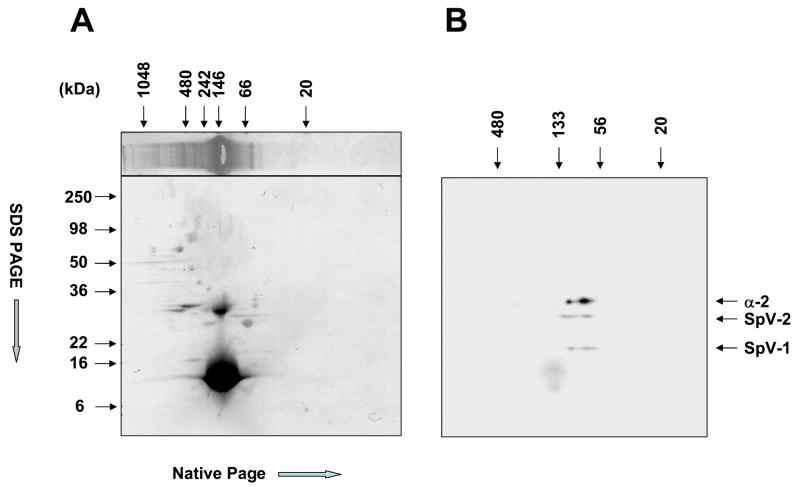
Two-dimensional (2D) electrophoresis of SpV-1/SpV-2 co-translation was performed to confirm the presence of SpV-1 and SpV-2 in the higher order complexes separated in native gels. The first dimension, native-PAGE, was followed by SDS-PAGE in the second dimension (Figure 7a). The Western blot of the 2D gel shows that the second dimension (SDS-PAGE) clearly separated SpV-1, SpV-2 and endogenous Alpha-2, confirming the presence of SpV-1 and SpV-2 proteins in complexes ranging from ~55kDa to 133 kDa (Figure 7b). This range overlaps but is not identical with the endogenous Alpha-2 and would include both dimeric and hetero-trimeric forms of canonical Paf-ah-Ib. The upper native-PAGE size range of SpV-1 is similar to Alpha-2; however, the lower range of SpV-1 is smaller than that of Alpha-2 or SpV-2. The SpV-2 native-PAGE size distribution (first dimension) has a component that is larger than the SpV-1 and endogenous Alpha-2 complexes (Figure 7b). The diffuse circular stained region of the immunostain (Figure 7B) running 16kDa and smaller in the second dimension is nonspecific chemiluminescent staining of the massive hemoglobin band from the reticulocyte lysate (Figure 7A).
Figure 7.
Two dimensional electrophoresis of unpurified SpV-1/SpV-2 rabbit reticulocyte lysate co-translation products. Panel A, upper horizontal lane is a representative lane from the same gel as used for the 2D analysis showing a Coomassie blue protein stain of the first dimension (Native-PAGE). The lower panel is the naphthol blue black protein stain of the completed 2D separation transfer. Panel B is the Pafah1b2 immunostain using chemiluminescent substrate of the 2D western blot showing the Alpha-2, SpV-2 and SpV-1 monomers.
Discussion
Although five new splice variants were found, they encode only two novel proteins exemplified by SpV-1 and SpV-2. These new proteins have identity with Alpha-2 for the first 137 amino acids, varying only in their carboxy termini (Figure 4). The variants replace exon 6 encoded amino acids (Alpha-2138–229) which abolishes the catalytic triad of the canonical Alpha-2 subunit by removing two key residues, Asp193 and His196 (ref 11, but with Alpha-2 numbering). Replacement of amino acids 138 to 229 of Alpha-2 also removes several residues involved in dimerization; however, the majority of the residues implicated in dimer formation (11) are encoded by exons 2–5 (amino acids 1–137) and are retained in SpV-1 and SpV-2. In-vitro translation experiments show that the replacement of Alpha-2 carboxy terminal amino acids in SpV-1 results in an association that is consistent with SpV-1 homo-dimer formation. In native PAGE experiments, in-vitro translated SpV-1 and SpV-2 were seen to form higher order interactions coincident with but not identical to the range seen for Alpha-2, raising the possibility that in addition to forming complexes with themselves or with canonical alpha subunits these novel subunits may be forming complexes with non-Paf-ah related proteins as well.
Variation in canonical alpha subunit composition of the paf-ah dimer has been shown to affect substrate specificity and catalytic activity. PAF was most efficiently hydrolyzed by the Alpha-2/Alpha-2 homodimer (12). In the Alpha-1/Alpha-2 hetero-dimer, the Alpha-2 subunit is thought to be inactive, leaving catalysis to the Alpha-1 subunit (22, 23). In such a scenario where the active sites of the alpha subunits in the paf-ah dimer are not required to work in concert, pairing one of the splice variants (SpV-1 or SpV-2) with a canonical catalytic alpha subunit (Alpha-1 or Alpha-2) could imbue the SpV/alpha heterodimer with altered substrate specificity, catalytic activity, or regulation by phosphorylation. It is also possible that the splice variants function independently from their catalytic cohort and complex with other proteins to play a separate role in cellular function. Alternatively, dimerization could occur in a combinatorial fashion assembling homo and hetero dimers from all forms of the protein producing a range of catalytic activities and regulatory possibilities.
The novel sequences of the new splice variants are not homologous to known proteins but do contain several predicted phosphorylation sites for kinases that have substantial roles in cell cycle regulation and cell division. This link to cell division bears an interesting parallel to the Beta subunit of Paf-ah-Ib (LIS1) which, in addition to its regulatory role in Paf-ah-Ib, is involved in microtubule associated processes in cell division (24). The association of LIS1 with microtubule associated proteins is reported to be regulated by phosphorylation (25), and a testis specific splice variant as well as polyadenylation variants have been reported for LIS1 mRNA in mice (26). Although phosphorylation of catalytic Paf-ah alpha subunits have not been reported, other members of the phospholipase A2 family, such as cPLA2 alpha, have been shown to be regulated by phosphorylation (27).
Does Addition of Exon AE-D Mark Alpha-2 Splice Variants for Degradation?
Splice variants SpV-3 and SPV-4 have an additional exon (AE-D) spliced to AE-C that creates a new 3′ end for the mRNA but adds no new coding sequence. AE-D is spliced 119 or 142 nucleotides downstream from the AE-C stop codon. This exon organization in which a splice junction is located more than 55 nucleotides downstream from the stop codon is consistent with transcripts involved in nonsense mediated mRNA decay (NMD) (28, 29). The inclusion of exon AE-D in Pafah1b2 transcripts may add NMD as an additional mechanism to modulate the level of Pafah1b2 mRNA within the cell.
A Testis Specific Splice Variant
SpV-2 encodes a testis specific protein isoform of Pafah1b2. Knockout mice models have shown that a homozygous knockout of the Pafah1b2 gene leads to infertility in male mice but does not affect fertility in females. Male infertility was due to the impairment of spermatogenesis at several stages of development. Homozygous knockout of the Alpha-1 subunit does not impair fertility or spermatogenesis (14, 30). The possibility arises that the modified protein encoded by SpV-2 either alone or in a complex with SpV-1 or canonical Paf-ah-Ib subunits, imparts the characteristics necessary for Pafah1b2’s specialized roles in spermatogenesis.
It has been suggested that Paf-ah-Ib’s activity in spermatogenesis, apoptosis, and neuronal migration may not rest entirely in its catalytic ability to reduce PAF levels but that it may act as a signaling complex in conjunction with other proteins (4, 5, 14). The Alpha-2 isoforms described herein may introduce levels of regulation and interaction beyond Pafah1b2’s catalytic activity imparting additional, as yet uncharacterized, links to intracellular signaling pathways in testis and other tissues.
Acknowledgments
This study was supported by NHLBI grant PHS HL46703. The Vermont Cancer Center DNA Analysis Facility for processing cDNA sequencing reactions, the technical assistance of Julia Valliere, and Dr. C.J. Palmer for her comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zimmerman GA, McIntyre TM, Prescott SM, Stafforini DM. The platelet-activating factor signaling system and its regulators in syndromes of inflammation and thrombosis. Crit Care Med. 2002;30(5 Suppl):S294–301. doi: 10.1097/00003246-200205001-00020. [DOI] [PubMed] [Google Scholar]
- 2.Ishii S, Shimizu T. Platelet-activating factor (PAF) receptor and genetically engineered PAF receptor mutant mice. Prog Lipid Res. 2000;39(1):41–82. doi: 10.1016/s0163-7827(99)00016-8. [DOI] [PubMed] [Google Scholar]
- 3.Liu LR, Xia SH. Role of platelet-activating factor in the pathogenesis of acute pancreatitis. World J Gastroenterol. 2006;12(4):539–45. doi: 10.3748/wjg.v12.i4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokuoka SM, Ishii S, Kawamura N, Satoh M, Shimada A, Sasaki S, Hirotsune S, Wynshaw-Boris A, Shimizu T. Involvement of platelet-activating factor and LIS1 in neuronal migration. Eur J Neurosci. 2003;18(3):563–70. doi: 10.1046/j.1460-9568.2003.02778.x. [DOI] [PubMed] [Google Scholar]
- 5.Bonin F, Ryan SD, Migahed L, Mo F, Lallier J, Franks DJ, Arai H, Bennett SA. Anti-apoptotic actions of the platelet-activating factor acetylhydrolase I alpha2 catalytic subunit. J Biol Chem. 2004;279(50):52425–36. doi: 10.1074/jbc.M410967200. [DOI] [PubMed] [Google Scholar]
- 6.Marcheselli VL, Rossowska MJ, Domingo MT, Braquet P, Bazan NG. Distinct platelet-activating factor binding sites in synaptic endings and in intracellular membranes of rat cerebral cortex. J Biol Chem. 1990;5;265(16):9140–5. [PubMed] [Google Scholar]
- 7.Bazan NG, Allan G. Platelet-activating factor in the modulation of excitatory amino acid neurotransmitter release and of gene expression. J Lipid Mediat Cell Signal. 1996;14(1–3):321–30. doi: 10.1016/0929-7855(96)00541-x. [DOI] [PubMed] [Google Scholar]
- 8.Stafforini DM, McIntyre TM, Zimmerman GA, Prescott SM. Platelet-activating factor acetylhydrolases. J Biol Chem. 1997;272(29):17895–8. doi: 10.1074/jbc.272.29.17895. [DOI] [PubMed] [Google Scholar]
- 9.Arai H. Platelet-activating factor acetylhydrolase. Prostaglandins Other Lipid Mediat. 2002;68–69:83–94. doi: 10.1016/s0090-6980(02)00023-0. [DOI] [PubMed] [Google Scholar]
- 10.Derewenda ZS, Derewenda U. The structure and function of platelet-activating factor acetylhydrolases. Cell Mol Life Sci. 1998;54(5):446–55. doi: 10.1007/s000180050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho YS, Swenson L, Derewenda U, Serre L, Wei Y, Dauter Z, Hattori M, Adachi T, Aoki J, Arai H, Inoue K, Derewenda ZS. Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer. Nature. 1997;385(6611):89–93. doi: 10.1038/385089a0. [DOI] [PubMed] [Google Scholar]
- 12.Manya H, Aoki J, Kato H, Ishii J, Hino S, Arai H, Inoue K. Biochemical characterization of various catalytic complexes of the brain platelet-activating factor acetylhydrolase. J Biol Chem. 1999;274(45):31827–32. doi: 10.1074/jbc.274.45.31827. [DOI] [PubMed] [Google Scholar]
- 13.Adachi H, Tsujimoto M, Hattori M, Arai H, Inoue K. Differential tissue distribution of the beta-and gamma-subunits of human cytosolic platelet-activating factor acetylhydrolase (isoform I. Biochem Biophys Res Commun. 1997;233(1):10–3. doi: 10.1006/bbrc.1997.6383. [DOI] [PubMed] [Google Scholar]
- 14.Yan W, Assadi AH, Wynshaw-Boris A, Eichele G, Matzuk MM, Clark GD. Previously uncharacterized roles of platelet-activating factor acetylhydrolase 1b complex in mouse spermatogenesis. Proc Natl Acad Sci U S A. 2003;100(12):7189–94. doi: 10.1073/pnas.1236145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gene seq reference. hg17_knownGene_NM_002572 range = chr11:116520250–116544024). May 2004 human reference sequence is based on NCBI Build 35 from the International Human Genome Sequencing Consortium.
- 16.Strausberg RL, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–903. doi: 10.1073/pnas.242603899. Epub 2002 Dec 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UCSC human genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
- 18.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puntervoll P, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31(13):3625–30. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–62. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 21.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31(5):1375–86. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hattori M, Adachi H, Aoki J, Tsujimoto M, Arai H, Inoue K. Cloning and expression of a cDNA encoding the beta-subunit (30-kDa subunit) of bovine brain platelet-activating factor acetylhydrolase. J Biol Chem. 1995;270(52):31345–52. doi: 10.1074/jbc.270.52.31345. [DOI] [PubMed] [Google Scholar]
- 23.Sheffield PJ, McMullen TW, Li J, Ho YS, Garrard SM, Derewenda U, Derewenda ZS. Preparation and crystal structure of the recombinant alpha(1)/alpha(2) catalytic heterodimer of bovine brain platelet-activating factor acetylhydrolase Ib. Protein Eng. 2001;14(7):513–9. doi: 10.1093/protein/14.7.513. [DOI] [PubMed] [Google Scholar]
- 24.Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2(11):784–91. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 25.Sapir T, Cahana A, Seger R, Nekhai S, Reiner O. LIS1 is a microtubule-associated phosphoprotein. Eur J Biochem. 1999;265(1):181–8. doi: 10.1046/j.1432-1327.1999.00711.x. [DOI] [PubMed] [Google Scholar]
- 26.Peterfy M, Gyuris T, Grosshans D, Cuaresma CC, Takacs L. Cloning and characterization of cDNAs and the gene encoding the mouse platelet-activating factor acetylhydrolase Ib alpha subunit/lissencephaly-1 protein. Genomics. 1998;47(2):200–6. doi: 10.1006/geno.1997.5121. [DOI] [PubMed] [Google Scholar]
- 27.Kudo I, Murakami M. Phospholipase A2 enzymes. Prostaglandins Other Lipid Mediat. 2002;68–69:3–58. doi: 10.1016/s0090-6980(02)00020-5. [DOI] [PubMed] [Google Scholar]
- 28.Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118(Pt 9):1773–6. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 29.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100(1):189–92. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koizumi H, Yamaguchi N, Hattori M, Ishikawa TO, Aoki J, Taketo MM, Inoue K, Arai H. Targeted disruption of intracellular type I platelet activating factor-acetylhydrolase catalytic subunits causes severe impairment in spermatogenesis. J Biol Chem. 2003;278(14):12489–94. doi: 10.1074/jbc.M211836200. [DOI] [PubMed] [Google Scholar]