Abstract
A rickettsial bacterium in the genus Wolbachia is the cause of a unidirectional reproductive incompatibility observed between two major beetle pests of maize, the western corn rootworm, Diabrotica virgifera virgifera, and the Mexican corn rootworm, D. v. zeae. These subspecies are allopatric except for two known regions of sympatry in Texas and Mexico. We demonstrate that populations of D. v. virgifera, with the exception of two populations in southern Arizona, are infected with a strain of Wolbachia. Populations of D. v. zeae are not infected. Treatment of D. v. virgifera with tetracycline eliminated the Wolbachia and removed the reproductive incompatibility. Similar patterns of reproductive incompatibility exist among taxa of the cricket genus Gryllus. Gryllus assimilis, G. integer, G. ovisopis, G. pennsylvanicus, and G. rubens are infected with Wolbachia whereas G. firmus is usually not. Populations of G. rubens and G. ovisopis carry the same Wolbachia strain, which is distinct from that of G. integer. G. pennsylvanicus is infected with two Wolbachia strains, that found in G. rubens and one unique to G. pennsylvanicus. Moreover, a proportion of G. pennsylvanicus individuals harbors both strains. Wolbachia may have influenced speciation in some members of the genus Gryllus by affecting the degree of hybridization between species. Given that Wolbachia infections are relatively common in insects, it is likely that other insect hybrid zones may be influenced by infections with Wolbachia.
Rickettsial bacteria of the genus Wolbachia have been identified as intracellular parasites in several taxa of arthropods. Wolbachia induce cytoplasmic incompatibility in the majority of species infected (1, 2). This phenomenon is expressed when an infected male mates with a female that is not infected, when male and female are both infected with two different Wolbachia strains, or when the male is infected with two strains and the female is infected with a single strain of Wolbachia. Results from such crosses vary depending on the strain of bacterium and insect species infected and can range from severe incompatibility resulting in very few to no offspring (2), a medium level of incompatibility (3), to no incompatibility (4, 5). Wolbachia also have been documented to trigger parthenogenesis in some hymenopterans (6, 7) as well as feminization in several species of isopods (8, 9). Little is known as to the precise mechanism by which Wolbachia bacteria induce these phenomena in their respective hosts (10, 11). Wolbachia also have been implicated in the speciation of some parasitic wasps in the genus Nasonia (12). Here we provide evidence that Wolbachia infections are common in adult populations of the chrysomelid beetle Diabrotica virgifera virgifera Le Conte and several cricket species in the genus Gryllus and that these infections may serve as a reproductive barrier between sympatric populations that are currently designated as subspecies and species.
D. v. virgifera and D. v. zeae Krysan and Smith are major pests of maize in North America and northern Mexico. Character indices using scutellar color and elytral maculation patterns are used to distinguish the two subspecies (13). Krysan et al. (13) characterized beetles from Cuernavaca, Morelos, Mexico, as D. v. zeae and beetles from Union County, NM, as D. v. virgifera. Beetles from near Finney and Edmonson in Hale County, TX, and Registro, Durango, Mexico, were intermediate and could not be categorized as either, suggesting the presence of hybrid zones. When D. v. virgifera males from South Dakota were crossed with D. v. zeae females from central Mexico, <2% of the eggs hatched compared with the reciprocal and intra-subspecific crosses, where >50% of the eggs hatched (14). Rickettsial bacteria have been observed, using electron microscopy, in the testes and cells of the spermatheca of adult D. v. virgifera (15). Here we report that the infection is an α-Proteobacterium in the genus Wolbachia, that it causes cytoplasmic incompatibility, and that it occurs in D. v. virgifera populations throughout the Corn Belt.
Reproductive incompatibilities also have been described between several members of the cricket genus Gryllus (16–19). We focused on six closely related species found in the United States and Canada; G. pennsylvanicus, G. firmus, G. integer, G. ovisopis, G. rubens, and G. assimilis (Fig. 3b). Unidirectional mating incompatibility has been shown between G. pennsylvanicus males and G. firmus females derived from populations in the eastern United States (19). A hybrid zone has been described between these two species which appears to follow the Blue Ridge and Appalachian mountains in Virginia, Pennsylvania, New Jersey, and New York (20, 21). Other incompatibilities have been recorded within the genus Gryllus: few eggs hatched when G. rubens and G. assimilis were reciprocally crossed (22), and similar results were obtained when crossing G. rubens and G. pennsylvanicus (22). Bidirectional incompatibility also has been reported between populations of G. rubens and G. integer (17, 18) whereas unidirectional incompatibility has been shown between G. ovisopis males and G. firmus females (T. Walker, personal communication). Werren et al. (23) reported that G. pennsylvanicus is infected with Wolbachia. We confirmed this observation and found that G. assimilis, G. integer, G. ovisopis, and G. rubens also are infected whereas G. firmus is usually not. We determined that single and double infections with Wolbachia are common in the species of this genus and propose that they may explain the incompatibilities reported and play a role in the speciation of some of its members.
Figure 3.
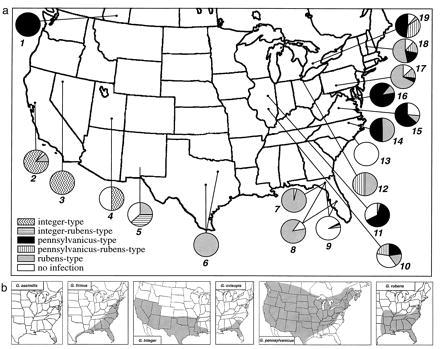
(a) Collection sites of Gryllus species and distribution of Wolbachia strains. Listed as: site number, species of Gryllus, locality, [number of crickets tested (strain type of Wolbachia identified in positives: P, pennsylvanicus; R, rubens; I, integer; P/R, pennsylvanicus/rubens; I/R, integer/rubens)]. 1, G. pennsylvanicus, Lethbridge, Alberta, Canada [3 (3 P)]; 2, G. integer, Davis, CA [15 (12I, 1R); 3, G. integer, Humboldt, County, NV [2 (2I)]; 4, G. integer, Wayne, County, UT [2 (1I)]; 5, G. integer, Las Cruces, NM [8 (2R, 3R/I)]; 6, G. integer, Dallas and Austin, TX [21 (21R)]; 7, G. ovisopis, Gainesville, FL [13 (12R)]; 8, G. rubens, Gainesville and Lake Placid, FL [11 (9R)]; 9, G. firmus, Gainesville and Lake Placid, FL [36 (2R, 1P)]; 10, G. nr. pennsylvanicus, Farina, IL [17 (4P, 3R, 4P/R)]; 11, G. nr. pennsylvanicus, Savoy, St. Joseph, Mahomet, and Urbana, IL [56 (38P, 1R)]; 12, G. nr. pennsylvanicus, Ann Arbor, MI [2 (1R, 1P/R)]; 13, G. firmus, Fort Fischer, NC [3]; 14, G. nr. pennsylvanicus, Winston–Salem, NC [2 (1P, 1R)]; 15, G. nr. pennsylvanicus, Charlottesville and Waynesboro, VA, and Augusta and Nelson Counties, VA [19 (12P, 2R)]; 16, G. nr. pennsylvanicus, Fairfax, VA [9 (7P, 1R, 1P/R)]; 17, G. nr. pennsylvanicus, Fayette, County, PA [15 (1P, 10R, 2P/R)]; 18, G. nr. pennsylvanicus, Norwich, VT [42 (7P, 22R, 7P/R)]; and 19, G. nr. pennsylvanicus, London, Ontario, Canada [21 (10P, 1R, 8P/R)]. (b) Maps showing the range of G. assimilis, G. firmus, G. integer, G. pennsylvanicus, and G. rubens in the United States and Canada (maps are courtesy of T. Walker; distribution of G. integer, D. Weissman personal communication).
MATERIALS AND METHODS
Insects.
A D. v. virgifera nondiapause strain derived from a South Dakota population in the early 1970s was used in the crosses. These beetles tested positive for Wolbachia. A Wolbachia-free colony of D. v. virgifera nondiapause strain was developed by feeding adults an artificial diet with 0.3% tetracycline for two generations. Subsequent generations were fed the same diet without tetracycline. This colony tested negative for Wolbachia at generations 2, 3, 5, and 7; individuals from the 8th generation were used for crosses. A D. v. zeae colony was initiated from a field collection near Stephenville, TX. D. v. zeae adults from the original collection and the second generation tested negative for Wolbachia. Second generation adults were used for crosses. To test for presence of infection in D. v. virgifera and D. v. zeae, we collected field populations from Guelph, Ontario, Canada; Freeville, NY; University Park, PA; Louisville, KY; East Lansing, MI; West Lafayette, IN; Urbana, IL; Madison, WI; Lamberton, MN; Ames, IA; Brookings, SD; Mead, NE; Fort Collins, CO; Moriarty, NM; Duncan and Willcox, AZ; three nearby sites: (i) 4 miles northeast of Edmonson, Swisher, County, TX; (ii) 6 miles north of Edmonson, Swisher, County, TX; (iii) 5 miles SE of Hart, Hale County, TX; Stephenville, TX; and Uvalde, TX.
Crosses.
Crosses with Diabrotica were made using teneral virgins kept as pairs in 300-cc cages at 25°C and 14:10 (light:dark). Eggs from each mated pair were collected in soil weekly for 7 weeks. A sample (total egg collection up to 100 eggs) of the weekly egg collection from each pair was plated on moist soil and maintained at 25°C. Eggs were monitored for up to 10 months until hatching was complete for all crosses.
Testing for, and Analysis of, Wolbachia Strains.
DNA was extracted using a phenol/chloroform procedure. Insects were screened for infection using the 99F and 994R 16S rRNA primers specific for Wolbachia (24). To ascertain the identity of the bacteria, we sequenced 1400 bp at the 5′ end of the 16S rRNA gene for all species identified as infected by PCR. These 1400 bp were sequenced from two overlapping PCR fragments obtained with primer pairs 21F and 994R and 99F and 1492R (24). A 956-bp segment of the ftsZ gene, whose product is involved in the septation of bacterial cells, also was sequenced for all species listed. For the D. v. virgifera specimens, this segment was obtained using primers ftsZfl and ftsZrl (23), whereas for the Gryllus specimens we used ftsZBf and ftsZBr primers specific for the B group Wolbachia (23). PCR products were isolated on a 1% NuSieve gels (FMC), purified using the Elu-quick DNA purification kit (Schleicher & Schuell), and sequenced directly in both directions. The 16S rRNA and ftsZ gene sequences for D. v. virgifera, G. assimilis, G. integer, G. ovisopis, G. pennsylvanicus, and G. rubens have been deposited in the GenBank database (accession nos. U83090–U83108 and AF011269–AF011271).
To identify the strains of Wolbachia, we sequenced a 1400-bp fragment of the 16S rRNA gene for one individual of: G. assimilis (T. Zera, colony); G. rubens and G. ovisopis, FL; G. integer, TX; G. integer, CA; G. integer, NV; G. integer, UT; G. pennsylvanicus, IL; D. v. virgifera, IL; D. v. virgifera nondiapause strain, SD; as well as a 956-bp fragment of the cell division ftsZ gene of G. assimilis (T. Zera, colony); G. rubens (T. Walker, colony); G. rubens and G. ovisopis, FL; G. integer, CA; G. integer, NV; two individuals of G. integer, NM; G. pennsylvanicus, IL; G. pennsylvanicus, VA; and D. v. virgifera from NM, IL, and VA. Once the DNA sequence of the various strains of Wolbachia was determined, strain identification in subsequent specimens was determined by restriction enzyme digest of the ftsZ gene. For D. v. virgifera, we used the restriction enzyme DraI. Wolbachia strains in the Gryllus species were differentiated with the following diagnostic restriction enzymes: Specimens infected with the pennsylvanicus-type strain of Wolbachia were distinguished using NsiI. Specimens infected with the rubens- and integer-type strain of Wolbachia were identified with BpmI and AvaII, respectively. For three G. assimilis individuals, the assimilis-type strain was identified by sequencing, in both directions, a region of the 16S rRNA gene using primers 261F and 517R (courtesy of C. Woese) to discern the presence of a thymine at Escherichia coli position 377 rather than a guanine common to all other known 16S rRNA gene Wolbachia sequences. Doubly infected Gryllus were at first identified by digestion of PCR products; the resulting uncut product was gel-purified and sequenced directly in both directions. Subsequent detections of double infections were determined by multiple restriction enzyme digests of PCR products (Fig. 2).
Figure 2.
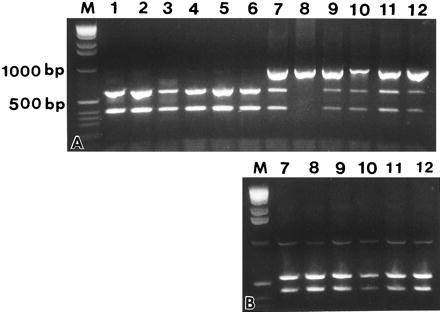
Restriction enzyme digest of PCR fragments with primers ftsZBf and ftsZBr. (a) Digests with BpmI. (b) Undigested products from a, purified and cut with NsiI. M, BRL 1-kb ladder; 1, 2, 8–12 G. nr. pennsylvanicus, Farina, IL; 6, 7 G. nr. pennsylvanicus, Ann Arbor, MI; 4, 5, G. nr. pennsylvanicus, Norwich, VT; and 3, G. firmus, Gainesville, FL.
DNA sequences of the ftsZ gene were first conceptually translated using dna strider (Christian Marck, Cedex, France), and then the amino acid sequences were aligned using clustal w, version 1.5 (25). DNA sequences were aligned following the alignment of amino acid sequences. The phylogenetic analysis was performed using only DNA sequences. These included ftsZ Wolbachia sequences in GenBank with the exception of the reported A strain of Tribolium confusum, which has been shown not to occur in this species (26). Maximum parsimony analysis was performed using paup, Ver. 3.1.1 (27). Because of the large number of taxa, only heuristic searches were performed. One hundred replications of each search were performed with random addition of taxa and tree bisection-reconnection branch swapping. Because no reasonable outgroup is available for the ftsZ gene, the resulting trees were midpoint rooted.
Geographic Distribution of Wolbachia Infection in Diabrotica and Gryllus Species.
A minimum of four individuals of D. v. virgifera and D. v. zeae were tested for infection using PCR and primers specific to the Wolbachia 16S rRNA gene for each of the geographic localities sampled. Within the hybrid zone in northern Texas, we sampled seven individuals at three sites.
Gryllus species tested and locations from which they originated are listed in Fig. 3. Specimens from Winston–Salem, NC; Nelson County, Augusta County, Waynesboro, and Charlottesville, VA, as well as some of the specimens from Fayette County, PA, are at the University of Michigan Museum. The remaining specimens are in the possession of R.G.
RESULTS AND DISCUSSION
Diabrotica is a New World genus of chrysomelid beetles that is primarily Neotropical in its distribution and whose origin is likely the region of Central or Tropical South America (28). Today its range encompasses most of the United States east of the Rockies and north of the 35th parallel, the states of Mexico northwest of Durango (excluding Baja, CA), southern areas of Ontario, Canada, and a recent introduction into eastern Europe (29). The distribution of D. v. zeae encompasses an area from Guatemala throughout central Mexico into the panhandle of Texas and parts of southern Oklahoma (28). Expanded maize production may also have facilitated the movement of D. v. zeae northward from Mexico, resulting in a region of secondary contact with D. v. virgifera in northern Texas, where the subspecies hybridize (13).
Adults of D. v. zeae sampled from Stephenville and Uvalde in southern Texas were not infected with Wolbachia whereas adults of D. v. virgifera from 15 sites within the United States and Canadian distribution of D. v. virgifera tested positive for Wolbachia, with the exception of two sites at Duncan and Willcox, AZ. Twenty-one adults from three sites within the north Texas hybrid zone also tested positive, including five specimens with index patterns within the range of variation of D. v. zeae (J. Krysan personal communication). Sequences of the ftsZ gene of Wolbachia from single individuals from Urbana, IL, Moriarty, NM, and Blacksburg, VA, were identical. In addition, digestion with DraI of the ftsZ fragment obtained with (i) general B-specific primers, (ii) ftsZf1, and (iii) ftsZr1 indicated that populations of D. v. virgifera from all sampled locations carry the same strain of Wolbachia. A phylogenetic analysis using the 16S rRNA gene placed this strain in a basal position with an unresolved relationship to other Wolbachia strains (results not shown). The ftsZ gene (23, 30) also identified the Wolbachia from D. v. virgifera as a basal strain and distinct from most other strains in the A group (Fig. 1). Given the consistency of infection and strain-type throughout the distribution of D. v. virgifera, as well as the rapid spread of this species in North America, we suggest that the infection occurred in the southwestern United States or northwestern Mexico before the eastward expansion of D. v. virgifera in the United States and Canada. The uninfected D. v. virgifera populations in southern Arizona may be the remnants of the original uninfected population or could represent isolated populations that have lost the infection.
Figure 1.
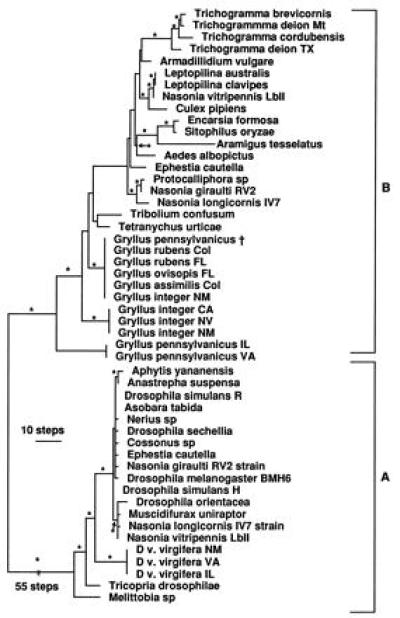
One of 36 equally most parsimonious trees estimated using a partial DNA sequence of the ftsZ gene. Asterisks indicate branches supported by all of the most parsimonious trees. The tree was midpoint rooted, had a length of 370 steps, a consistency index (excluding uninformative characters) of 0.64, and a retention index of 0.95. Designation A and B refer to groups identified by Werren et al. (23). †, Wolbachia sequence published in Werren et al. (23).
D. v. virgifera and D. v. zeae are morphologically very similar (13), with the exception of black elytral maculation that is distinctive in D. v. virgifera and confined to the humerus or is absent in D. v. zeae. The subspecies are not ecologically isolated because they are both found on maize; nor are they temporally isolated because they have the same diapause mechanism. Behavioral studies do not indicate any premating barriers to reproduction because both subspecies respond to the same sex pheromone and no discrimination was made during mating choice experiments (13). When crossed, the two subspecies were fertile in one direction, i.e., D. v. zeae males from central Mexico mated with D. v. virgifera females from South Dakota whereas the reciprocal cross produced very few to no viable offspring (14). To test whether the unidirectional incompatibility is caused by the Wolbachia infection identified in D. v. virgifera, we made reciprocal crosses from infected and tetracycline-treated D. v. virgifera. The results indicate that the strain of Wolbachia that infects D. v. virgifera is capable of inducing severe cytoplasmic incompatibility, with only 0.4% of 5627 eggs hatching when infected D. v. virgifera males were crossed with tetracycline-treated uninfected D. v. virgifera females (Table 1, cross d).
We also established a laboratory colony of D. v. zeae with specimens from Stephenville, TX, and confirmed that D. v. zeae females crossed with D. v. virgifera males produce very few to no offspring whereas the reciprocal cross results in a normal egg hatch (Table 1, crosses f and h) (14). When we repeated the crosses using tetracycline-treated D. v. virgifera males, the incompatibility no longer existed, and egg hatch rates were observed that were not significantly different from the controls (Table 1, cross g), confirming that the unidirectional mating incompatibility between these two subspecies is caused by the presence of Wolbachia in D. v. virgifera.
Similar reproductive incompatibilities also have been reported between some members of the genus Gryllus in North America. The genus Gryllus is distributed throughout most of the world with the exception of eastern Asia and the Pacific (31). Based on numbers of species, it is possible that the genus originated in the New World or Africa, where several genera similar to Gryllus have been recorded (32). In North America, the genus Gryllus has several closely related species, some of which are difficult to distinguish on the basis of morphological characters alone. At the beginning of the century, this group was regarded as a single species with a high degree of variability (33). Subsequent work mainly using calling songs discriminated among some of the species (34).
Several species in the genus Gryllus have been shown to be unidirectionally or bidirectionally incompatible. The unidirectional incompatibility between G. pennsylvanicus males and G. firmus females in the eastern United States (19) mirrors that of D. v. virgifera and D. v. zeae. We have found that, with the exception of G. firmus, specimens from all of the Gryllus species that we have studied are usually infected with Wolbachia, sometimes with more than one strain (Figs. 2 and 3). The incompatibilities reported between the various species in this genus may be explained by infection with this bacterium. Our study has shown the following patterns of infection. Populations of G. integer sampled from Humboldt County, NV, and Wayne County, UT, are infected with the same strain of Wolbachia bacteria, which we refer to as the “integer type.” The population sampled in Davis, CA, also was infected with the integer type, with the exception of one individual that harbored the same strain of Wolbachia as found in G. rubens, which we refer to as the “rubens type.” The population sampled in Las Cruces, NM, had two individuals that were infected with the rubens-type Wolbachia strain and three individuals that were doubly infected with the rubens- and integer-type strains. The G. integer populations in Austin and Dallas, TX, also harbored the same bacterial strain, which, on the basis of the 16S rRNA gene and the ftsZ gene, appears identical to that found in G. rubens from Lake Placid and Gainesville, FL. This rubens-type strain also occurs in specimens of G. ovisopis sampled from Gainesville. We could only obtain specimens of G. assimilis from the laboratory cultures of T. Walker and T. Zera; the latter culture was derived from T. Walker’s colony. Individuals from both cultures were infected with the same strain whose sequence for the ftsZ gene was identical to the rubens-type Wolbachia strain. The 16S rRNA gene fragment sequence indicates that the strain in this species has a transversion at E. coli position 380 from a guanine to a thymine, which in E. coli corresponds to the first position in the tetra loop GCAA. In the “assimilis-type” strain, the loop is comprised of TAAA. This transversion is not found in any other Wolbachia 16S rRNA sequence known to date.
The population of G. pennsylvanicus shows a more complex pattern of infection. Throughout most of the distribution that we have sampled, G. pennsylvanicus is polymorphic for infection type. A population sampled in Lethbridge, Alberta, Canada, showed the presence of a Wolbachia strain different from those found in G. rubens, G. assimilis, G. integer, and G. ovisopis. We refer to this strain as the “pennsylvanicus-type” strain. Of 39 infected specimens sampled from central Illinois, 38 harbored the pennsylvanicus-type strain whereas one specimen carried the rubens-type strain. A locality 150 km to the south, Farina, IL, showed a mixture of individuals that were infected with either the pennsylvanicus-type or rubens-type strain or with both. Of two specimens sampled from Ann Arbor, MI, one carried the rubens-type strain, and the other was doubly infected with the pennsylvanicus- and rubens-type strains. For the population sampled in London, Ontario, Canada, 215 km northeast of Ann Arbor, of 19 infected individuals, 10 carried the pennsylvanicus-type, one carried the rubens-type, and eight were doubly infected with the pennsylvanicus- and rubens-type strains (Fig. 3a). This polymorphic pattern continues in specimens of G. pennsylvanicus collected from Vermont and Pennsylvania. In Virginia and North Carolina, we detected only individuals that were infected with either the pennsylvanicus- or rubens-type strains. The population of G. pennsylvanicus in North America is a mixture of individuals that are infected with the rubens-type or pennsylvanicus-type as well as individuals that are doubly infected with both strains and individuals that are not infected (Fig. 3). Our results thus far indicate that, in populations of G. pennsylvanicus of eastern North America, the rubens-type infection is at a higher frequency in the north than in the south (Fig. 3a). Furthermore, we have found that, in most localities where both strains occur, doubly infected individuals are also present (Figs. 2 and 3a).
As a result of the “drive” associated with the spread of a Wolbachia infection (35), we propose that the genetic structure of G. pennsylvanicus populations we have sampled, excluding Lethbridge, Canada, have been affected by the presence of the rubens- and pennsylvanicus-type Wolbachia strains. Therefore, we shall refer to the G. pennsylvanicus samples collected in these regions as Gryllus near (nr.) pennsylvanicus to reflect the ambiguity of their taxonomic status imposed by the infection types found in these populations.
We have sampled G. firmus from Gainesville and the Archbold Biological Station in Lake Placid, FL, and from Fort Fischer, NC. Thirty-two of the 35 specimens sampled from these localities were uninfected. The three infected individuals came from Gainesville, FL. Two were found to carry the rubens-type, and the third carried the pennsylvanicus-type Wolbachia strain (Figs. 2 and 3a). This finding suggests that infection of these two strains of bacteria is moving into the populations of G. firmus, especially in light of the fact that G. pennsylvanicus is not known from Gainesville, FL.
Analysis of the relationships among the different strains we have examined of Wolbachia in the Gryllus species using the ftsZ gene indicates that the integer- and rubens-type strains form a monophyletic group and are not closely related to the pennsylvanicus-type strain, which is basal to all other B group Wolbachia strains known thus far. The strain of Wolbachia reported by Werren et al. (23) in G. pennsylvanicus is of the rubens-type (Fig. 2).
We propose that the infections in the above members of the genus Gryllus offer an explanation for the observed mating incompatibilities obtained by Smith and Cade (17), Cade and Tyshenko (18), and Harrison (19) and point to a possible role of Wolbachia in the speciation of some members of this genus. The bidirectional incompatibility observed between G. rubens and G. integer is likely to be the result of infection with the two different strains of Wolbachia found in these two species. The compatibility of G. rubens from Florida with G. integer from Texas (17, 18) can be explained because these populations carry the rubens-type bacteria. Our data lend support to the conclusion of Smith and Cade (17) that G. rubens and G. integer from Texas may be the same species, despite differences in their songs of a degree usually used to differentiate species.
Given that G. pennsylvanicus is infected with Wolbachia and G. firmus is usually not, crosses showing that G. pennsylvanicus males are incompatible with G. firmus females (19) can be interpreted as resulting from cytoplasmic incompatibility caused by Wolbachia. Our interpretation is further substantiated by the mitochondrial pattern observed in these two populations, namely that G. firmus and G. pennsylvanicus have distinct mitochondrial types by which these two populations have been identified; however, although G. pennsylvanicus mtDNA readily introgresses in the population of G. firmus, the reverse does not occur (36). This pattern would be observed if G. pennsylvanicus is infected with Wolbachia because the majority of individuals in the hybrid zone would be the result of progeny of infected females, which in this case would carry the G. pennsylvanicus mitochondrial type together with the maternally inherited Wolbachia infection.
The unidirectional incompatibility between G. ovisopis males, which carry the rubens-type bacteria, and G. firmus females (T. Walker, personal communication) also can be interpreted in light of this infection. The lack of progeny from crosses between males of G. pennsylvanicus and G. ovisopis with females of G. firmus indicate that these two strains might cause incompatibility. We know that G. ovisopis and G. rubens carry the same Wolbachia strain, so the likely cause of the bidirectional incompatibility observed between G. rubens and G. pennsylvanicus is the different Wolbachia strains they can harbor.
Using the ftsZ gene, we observed 39 bp differences between the pennsylvanicus- and rubens-type strains, 41 differences between the pennsylvanicus- and integer-type strains, and 22 differences between the rubens- and integer-type strains. For a comparison, in the same gene segment, there is only one base change between the bidirectionally incompatible Wolbachia strains found in Drosophila simulans Hawaii and D. simulans Riverside (23). Based on this evidence as well as on the incompatibilities observed in the matings between the respective Gryllus species, we predict that the rubens-, integer-, and pennsylvanicus-type strains are bidirectionally incompatible.
The hybrid zone between G. pennsylvanicus and G. firmus designated by Harrison and Arnold (20) is more complex than a species–species interaction. Some of the sites that we sampled along the Blue Ridge and Appalachian mountains show a patchy distribution of infection types within G. pennsylvanicus populations (Fig. 3a). Thus, in this region, possible interactions can occur between populations of uninfected G. firmus, G. nr. pennsylvanicus infected with either rubens- or pennsylvanicus-type Wolbachia and G. nr pennsylvanicus infected with both rubens- and pennsylvanicus-type Wolbachia strains. The Wolbachia infection in the Gryllus species mentioned and specifically the multiple infection types found in the G. nr. pennsylvanicus population indicate that restriction to gene flow can occur within a population because of differences in infection-type. The reproductive success of individuals in these populations will depend on their infection profile as well as that of the individuals with which they mate. Our concept of what constitutes a species in the genus Gryllus must be re-evaluated in light of the Wolbachia infections that they harbor.
Infections with Wolbachia can sweep through a population as a result of the advantage that infected females have over uninfected females (35). Given that G. pennsylvanicus and G. firmus have been shown to hybridize in the laboratory (19) and that there is evidence for hybridization in the field (21), the current distribution of G. firmus can be interpreted as a relict one, from a wider area through which a sweep of an infected population moved. An alternative explanation could be multiple infection sites with the same Wolbachia strain, but we have no information as to how Wolbachia bacteria are transmitted horizontally across species or the frequency with which they are acquired. Therefore, we choose the most parsimonious explanation, that the infection originated at one location and subsequently spread. Indeed, our data suggest that the Wolbachia infections have a geographic component and that the rubens-type originated in the south where it predominates (Fig. 3a). However, in the G. nr. pennsylvanicus population of eastern North America, the rubens-type infection is prevalent in populations sampled in the north. These localities are the farthest points from the populations of Gryllus uniformly infected with the rubens-type strain and far from any reported G. rubens populations (Fig. 3 a and b). We propose that the rubens-type infection moved through the population of Gryllus that existed in eastern North America before the eastward movement of the pennsylvanicus-type strain.
In the Gryllus populations, doubly infected individuals may be the result of selection pressure in a population in which two bacterial strains exist that are bidirectionally incompatible (Figs. 2 and 3a). In such a circumstance, individuals that are doubly infected would be selected for because, if both infections can survive in one cricket, the females would be compatible with males carrying either bacterial strain. A third infective sweep of doubly infected individuals could then ensue through the population because doubly infected females have an advantage over singly infected females, which could only reproduce successfully with males infected with the same Wolbachia strain. Multiple infections have been reported from a variety of insect taxa (37–39), and doubly infected males have been shown to be partially or completely incompatible with singly infected females in other insects (37–41). We predict that double infections could occur in localities where single infections with two bidirectionally incompatible strains of Wolbachia occur in the same or very closely related taxa.
In a screen of temperate insect taxa using PCR with specific 16S rRNA primers, we found that 16% of 615 insect taxa, from a wide variety of families and orders, were infected with Wolbachia (R.G. and H.M.R., unpublished data), and similar infection rates have been obtained for tropical insects (42). Given this frequency of infection, we predict that other insect hybrid zones will be found to be defined at least in part by Wolbachia infection and cytoplasmic incompatibility. Some genera of insects may be more prone to infection with Wolbachia as a consequence of their natural history. Speciation within these groups needs to be examined with respect to the role played by Wolbachia.
Table 1.
Comparisons of mean percent egg hatch ± SE for crosses using infected and tetracycline-treated D. v. virgifera and naturally uninfected D. v. zeae using the Mann–Whitney U test
| Female | Male | Cross | n | Eggs | Hatch, % | Comparison |
|---|---|---|---|---|---|---|
| D.v.v. | x D.v.v. | a | 10 | 4706 | 74.9 ± 5.5 | a vs. b NS |
| D.v.v. | x D.v.v.T | b | 10 | 5109 | 81.1 ± 3.4 | |
| D.v.v.T | x D.v.v.T | c | 10 | 5187 | 66.4 ± 6.2 | |
| D.v.v.T | x D.v.v. | d | 14 | 5627 | 0.4 ± 0.1 | d vs. c* |
| D.v.z. | x D.v.z. | e | 10 | 4608 | 83.4 ± 4.0 | |
| D.v.z. | x D.v.v. | f | 15 | 6433 | 0.3 ± 0.1 | f vs. e* |
| D.v.z. | x D.v.v.T | g | 15 | 6376 | 74.8 ± 6.5 | g vs. e NS |
| D.v.v. | x D.v.z. | h | 15 | 7735 | 82.0 ± 4.3 | h vs. a NS |
| D.v.v.T | x D.v.z. | i | 15 | 8447 | 75.5 ± 5.3 | i vs. c NS |
T, tetracycline-treated; D. v. v., D. v. virgifera; D. v. z., D. virgifera zeae; NS, not significant.
*P < 0.001.
Acknowledgments
We thank the following people: for Diabrotica specimens: L. Bledsoe, D. Calvin, L. Clark, S. Coleman, G. Cronholm, P. Davis, L. French, C. Ellis, M. Haas, M. Hoffman, A. Keaster, D. Landis, B. Lewis, L. Meinke, P. Morrison, G. Nardin, F. Peairs, J. Tollefson, L. Townsend, T. Turpin, J. Wedberg, and R. Youngman; for Gryllus specimens: R. Alexander, W. Cade, S. Carroll, R. Fialho, G. Fondufe, G. Hammond, L. Higgins, L. Heuser, D. Houchens, D. Lactin, D. Lampe, D. Lightfoot, D. Marshall, A. Murray, R. Ottens, B. Shipley, M. Upton, T. Zera, R. Walker, T. Walker, D. Weissman, A. Wild, J. Whistlecraft, and J. Watts; and for identification of insect specimens: R. Alexander, J. Krysan, D. Lightfoot, T. Walker, and D. Weissman. We thank T. Walker for allowing the use of distribution maps of Gryllus, as well as P. Patel, P. White, and K. Zumpano for technical assistance, and R. Fialho and L. Stevens for ftsZ primers. This work has benefited from discussions with R. Alexander, L. Higgins, J. Krysan, E. MacLeod, S. O’Neill, M. Turelli, T. Uzzell, T. Walker, and D. Weissman. W. Cade, J. Krysan, E. MacLeod, J. Nardi, F. Soto-Adames, and an anonymous reviewer made helpful comments on the manuscript. Support was provided by U.S. Department of Agriculture Grant 91-37302-6766, National Science Foundation Grant MCB 93-17586, and the University of Illinois Research Board.
Footnotes
References
- 1.Hsiao C, Hsiao T H. J Invertebr Pathol. 1985;45:244–246. [Google Scholar]
- 2.Wade M J, Stevens L. Science. 1985;227:527–528. doi: 10.1126/science.3966160. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman A A. Entomol Exp Appl. 1988;48:61–67. [Google Scholar]
- 4.Giordano R, O’Neill S L, Robertson H M. Genetics. 1995;140:1307–1317. doi: 10.1093/genetics/140.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann A A, Clancy D, Duncan J. Heredity. 1996;76:1–8. doi: 10.1038/hdy.1996.1. [DOI] [PubMed] [Google Scholar]
- 6.Zchori-Fein E, Faktor O, Zeidan M, Gottlieb Y, Czonek H, Rosen D. Insect Mol Biol. 1995;4:173–178. doi: 10.1111/j.1365-2583.1995.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 7.Stouthammer R, Pinto J D, Platner G R, Luck R F. Ann Entomol Soc Am. 1990;83:475–481. [Google Scholar]
- 8.Martin G, Juchault P, Legrand J J. C R Acad Sci Paris. 1973;276:2313–2316. [Google Scholar]
- 9.Juchault P, Frelon M, Bouchon D, Rigaud T. C R Acad Sci Paris. 1994;317:225–230. [Google Scholar]
- 10.Ryan S L, Saul G B. Mol Gen Genet. 1968;103:29–36. doi: 10.1007/BF00271154. [DOI] [PubMed] [Google Scholar]
- 11.Callaini G, Dallai R, Riparbelli G. J Cell Sci. 1997;110:271–280. doi: 10.1242/jcs.110.2.271. [DOI] [PubMed] [Google Scholar]
- 12.Breeuwer J A J, Werren H. Nature (London) 1990;346:558–560. doi: 10.1038/346558a0. [DOI] [PubMed] [Google Scholar]
- 13.Krysan J L, Smith R F, Branson T F, Guss P L. Ann Entomol Soc Am. 1980;73:123–130. [Google Scholar]
- 14.Krysan J L, Branson T F. J Hered. 1977;68:415–417. [Google Scholar]
- 15.Degrugillier M E, Degrugillier S S, Jackson J J. J Invertebr Pathol. 1991;57:50–58. [Google Scholar]
- 16.Alexander R D. Q Rev Biol. 1968;43:1–41. doi: 10.1086/405628. [DOI] [PubMed] [Google Scholar]
- 17.Smith C J, Cade W H. Can J Zool. 1987;65:2390–2394. [Google Scholar]
- 18.Cade W H, Tyshenko M G. Can J Zool. 1990;68:2697–2700. [Google Scholar]
- 19.Harrison R G. Evolution. 1983;37:245–251. doi: 10.1111/j.1558-5646.1983.tb05534.x. [DOI] [PubMed] [Google Scholar]
- 20.Harrison R G, Arnold J. Evolution. 1982;36:535–552. doi: 10.1111/j.1558-5646.1982.tb05075.x. [DOI] [PubMed] [Google Scholar]
- 21.Harrison R G, Rand D M, Wheeler W C. Mol Biol Evol. 1987;4:144–158. [Google Scholar]
- 22.Bigelow R S. Can J Zool. 1960;38:509–524. [Google Scholar]
- 23.Werren J H, Zhang W, Guo R L. Proc R Soc London Ser B. 1995;261:55–71. [Google Scholar]
- 24.O’Neill S L, Giordano R, Colbert A M E, Karr T L, Robertson H M. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;2:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fialho R F, Stevens L. Proc R Soc Lond Ser B. 1997;264:1065–1068. doi: 10.1098/rspb.1997.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. , Ver. 3.1.1. [Google Scholar]
- 28.Krysan J L, Smith R F. Entomography. 1987;5:375–484. [Google Scholar]
- 29.Maceljski M, Barcic J I. Fragm Phytom Herbol. 1993;21:173–183. [Google Scholar]
- 30.Holden P R, Brookfield J F Y, Jones P. Mol Gen Genet. 1993;240:213–220. doi: 10.1007/BF00277059. [DOI] [PubMed] [Google Scholar]
- 31.Alexander R D. Great Lakes Entomol. 1991;24:79–84. [Google Scholar]
- 32.Otte D, Cade W. Proc Nat Sci Philadelphia. 1984;136:98–122. [Google Scholar]
- 33.Rehn J A G, Hebard M. Proc Acad Nat Sci Philadelphia. 1915;67:293–322. [Google Scholar]
- 34.Alexander R D. Ann Entomol Soc Am. 1957;50:584–602. [Google Scholar]
- 35.Turelli M, Hoffmann A A. Nature (London) 1991;353:440–442. doi: 10.1038/353440a0. [DOI] [PubMed] [Google Scholar]
- 36.Harrison R G, Bogdanowicz S M. Evolution. 1997;51:493–505. doi: 10.1111/j.1558-5646.1997.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 37.Rousset F, Solignac M. Proc Natl Acad Sci USA. 1995;92:6389–6393. doi: 10.1073/pnas.92.14.6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinkins S P, Braig H R, O’Neill S L. Exp Parasitol. 1995;81:284–291. doi: 10.1006/expr.1995.1119. [DOI] [PubMed] [Google Scholar]
- 39.Perrot-Minnot M, Guo L R, Werren J H. Genetics. 1996;143:961–972. doi: 10.1093/genetics/143.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercot H, Llorente B, Jacques M, Atlan A, Montchamps-Moreau C. Genetics. 1995;141:1015–1023. doi: 10.1093/genetics/141.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinkins S P, Braig H R, O’Neill S L. Proc R Soc London Ser B. 1995;261:325–330. doi: 10.1098/rspb.1995.0154. [DOI] [PubMed] [Google Scholar]
- 42.Werren J H, Windsor D, Guo L R. Proc R Soc London Ser B. 1995;262:197–204. [Google Scholar]


