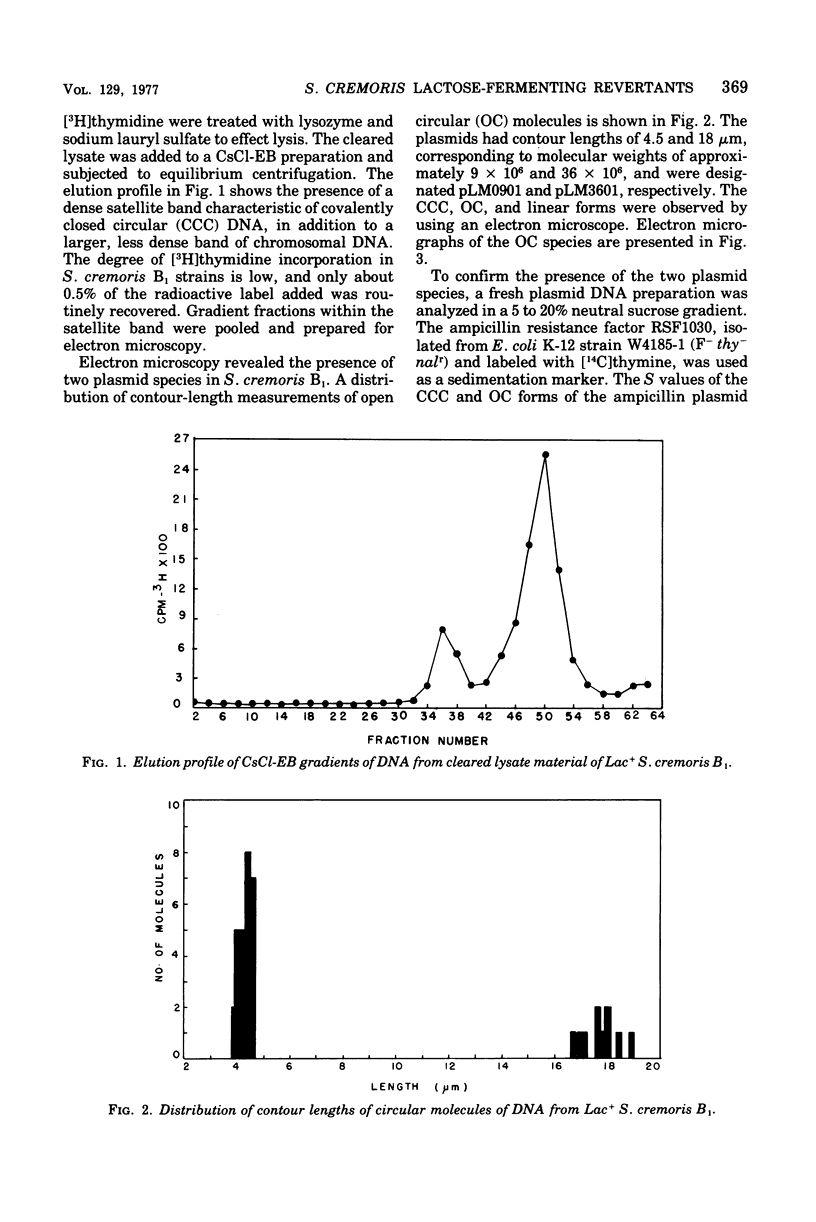
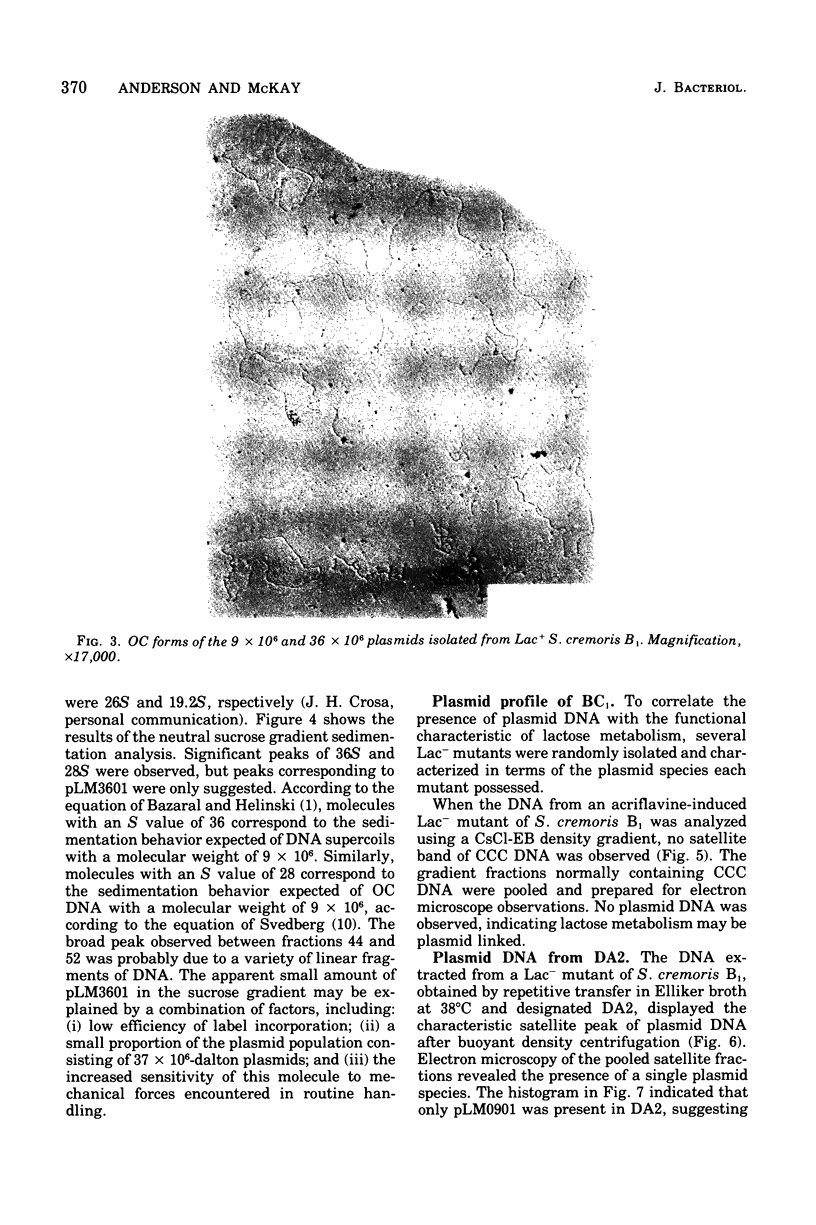
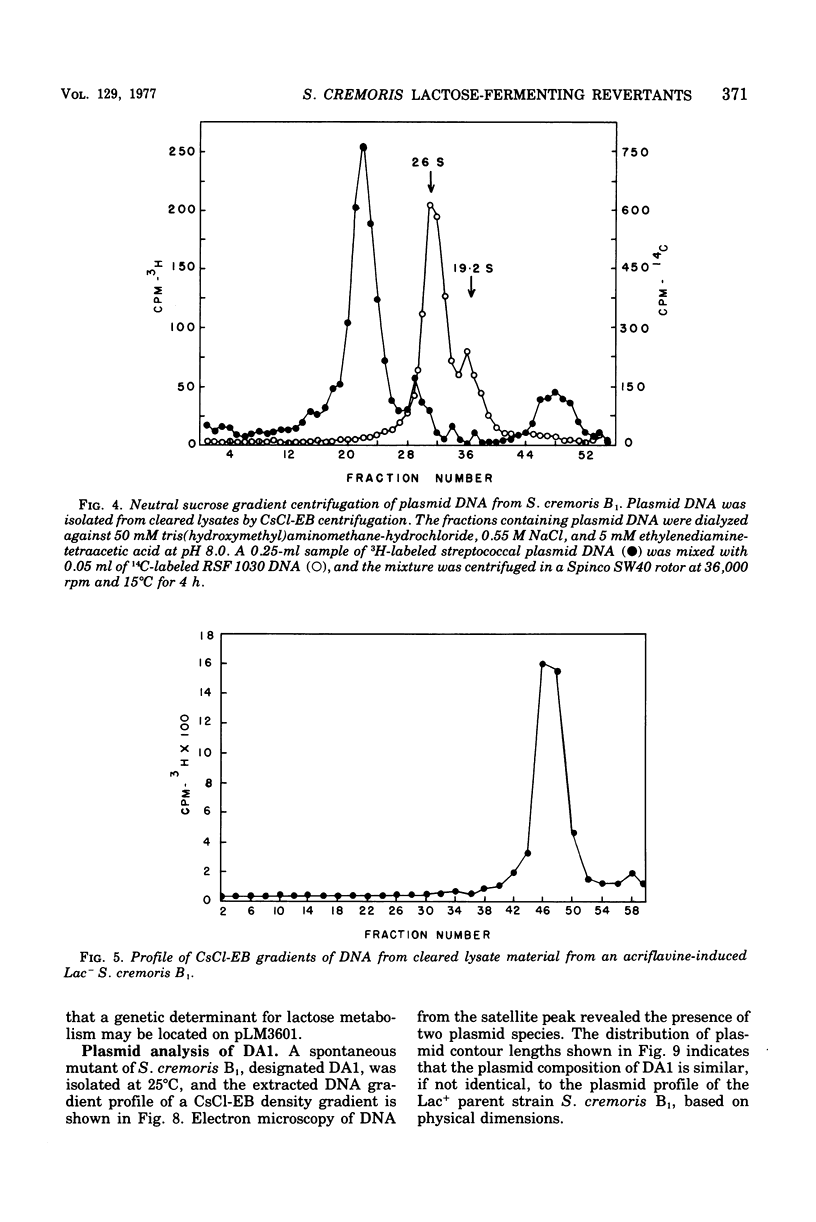
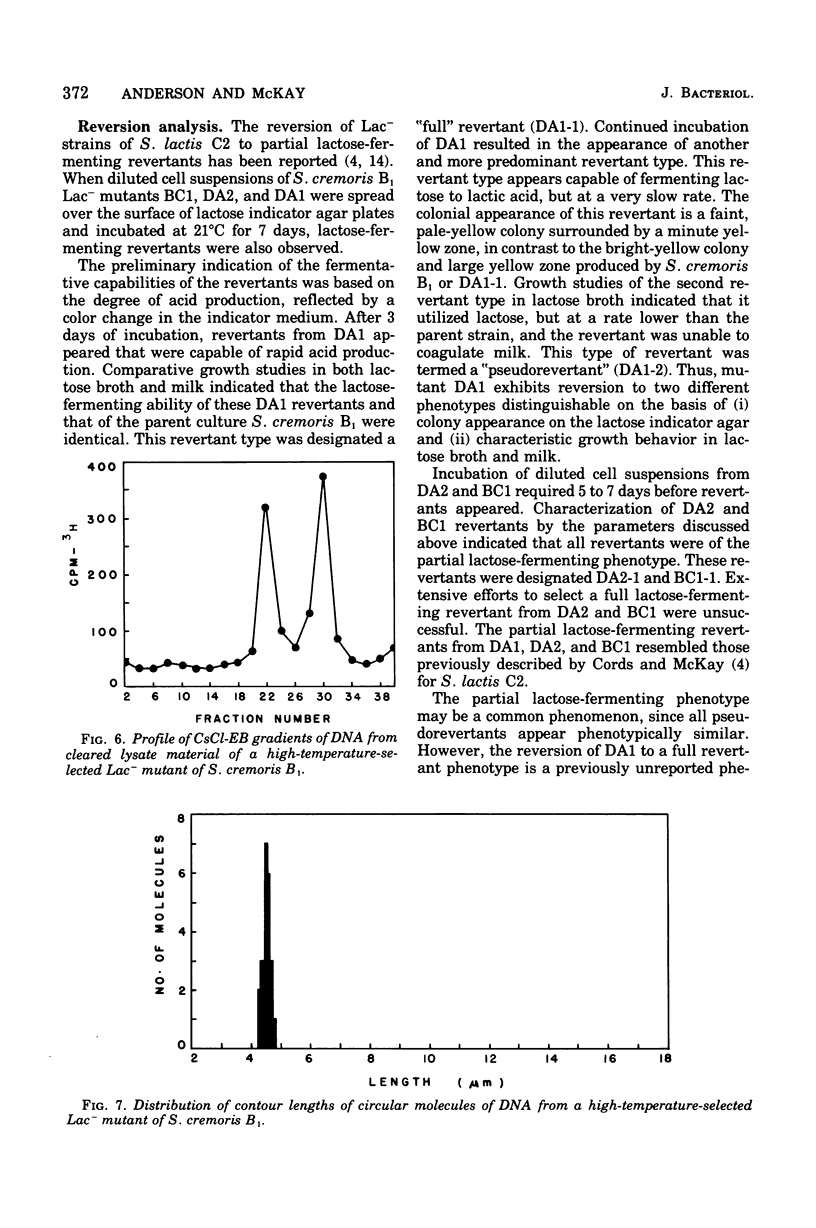
Abstract
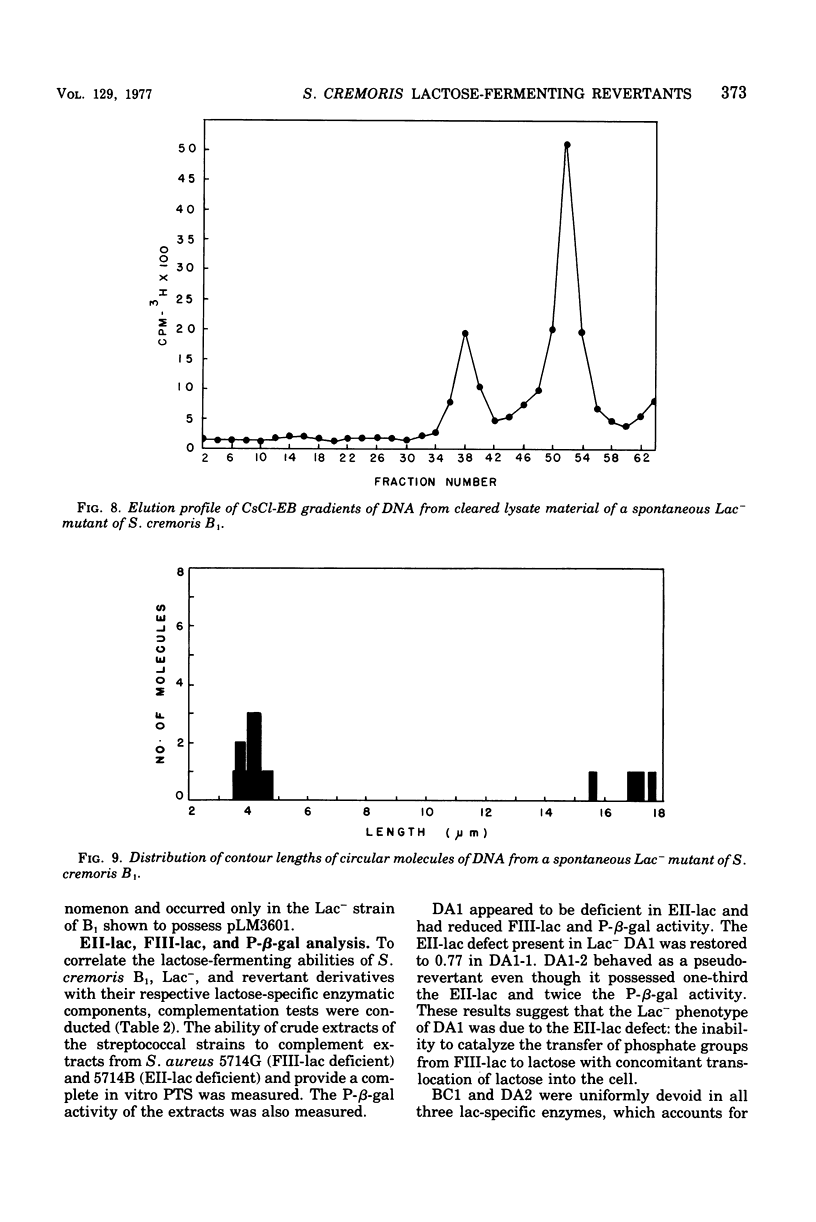
The unstable ability to metabolize lactose (lac) via the phosphoenolpyruvate-phosphotransferase system (PTS) was examined in Streptococcus cremoris B1. The presence of functional lactose-specific PTS enzymes was correlated with the presence of a distinct plasmid species. Characterization of deoxyribonucleic acid extracted from lactose-positive (Lac+) S. cremoris B1 revealed two plasmids having molecular weights of 9 X 10(6) and 36 X 10(6). An acriflavine (BC1)-induced, lactose-negative (Lac-) mutant possessed no plasmids and was devoid of all three lac-specific PTS enzymes. A Lac- mutant (DA2) isolated by growing at elevated temperatures only possessed the 9 X 10(6)-dalton plasmid and also lacked the lac PTS enzymes. A spontaneous Lac- mutant possessed both the 9 X 10(6)-and 36 X 10(6)-dalton plasmids. This mutant displayed FIII-lac and phospho-beta-D-galactosidase (P-beta-gal) activity but was deficient in EII-lac activity. The spontaneous Lac- strain reverted to both full and partial lactose-fermenting phenotypes having FIII-lac, EII-lac, and P-beta-gal activities. BC1 and DA2 Lac- mutants reverted only to the partial lactose-fermenting phenotype having P-beta-gal activity; EII-lac and FIII-lac activities were absent. The results indicate that the genetic determinants for EII-lac, FIII-lac, and P-beta-gal are located on the 36 X 10(6)-dalton plasmid in S. cremoris B1. Evidence for a second chromosomally associated P-beta-gal gene operating in the partial lactose-fermenting revertants is also presented.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazaral M., Helinski D. R. Characterization of multiple circular DNA forms of colicinogenic factor E-1 from Proteus mirabilis. Biochemistry. 1968 Oct;7(10):3513–3520. doi: 10.1021/bi00850a028. [DOI] [PubMed] [Google Scholar]
- Bissett D. L., Anderson R. L. Lactose and D-galactose metabolism in group N streptococci: presence of enzymes for both the D-galactose 1-phosphate and D-tagatose 6-phosphate pathways. J Bacteriol. 1974 Jan;117(1):318–320. doi: 10.1128/jb.117.1.318-320.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. H., Lengyel J. A., Langridge J. Evolution of a second gene for beta-galactosidase in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cords B. R., McKay L. L. Characterization of lactose-fermenting revertants from lactose-negative Streptococcus lactis C2 mutants. J Bacteriol. 1974 Sep;119(3):830–839. doi: 10.1128/jb.119.3.830-839.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cords B. R., McKay L. L., Guerry P. Extrachromosomal elements in group N streptococci. J Bacteriol. 1974 Mar;117(3):1149–1152. doi: 10.1128/jb.117.3.1149-1152.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H., Luttropp L. K., Falkow S. Nature of R-factor replication in the presence of chloramphenicol. Proc Natl Acad Sci U S A. 1975 Feb;72(2):654–658. doi: 10.1073/pnas.72.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou J. D., McKay L. L. Plasmids in Streptococcus lactis: evidence that lactose metabolism and proteinase activity are plasmid linked. Appl Environ Microbiol. 1976 Jul;32(1):38–44. doi: 10.1128/aem.32.1.38-44.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRSCH A. Growth and nisin production of a strain of Streptococcus lactis. J Gen Microbiol. 1951 Feb;5(1):208–221. doi: 10.1099/00221287-5-1-208. [DOI] [PubMed] [Google Scholar]
- Hengstenberg W., Penberthy W. K., Hill K. L., Morse M. L. Phosphotransferase system of Staphylococcus aureus: its requirement for the accumulation and metabolism of galactosides. J Bacteriol. 1969 Aug;99(2):383–388. doi: 10.1128/jb.99.2.383-388.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B., Clayton D. A., Vinograd J. Complex mitochondrial DNA. Cold Spring Harb Symp Quant Biol. 1968;33:435–442. doi: 10.1101/sqb.1968.033.01.050. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lang D. Molecular weights of coliphages and coliphage DNA. 3. Contour length and molecular weight of DNA from bacteriophages T4, T5 and T7, and from bovine papilloma virus. J Mol Biol. 1970 Dec 28;54(3):557–565. doi: 10.1016/0022-2836(70)90126-9. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Efstathiou J. D. Transductional evidence for plasmid linkage of lactose metabolism in streptococcus lactis C2. Appl Environ Microbiol. 1976 Jul;32(1):45–52. doi: 10.1128/aem.32.1.45-52.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Walter L. A., Sandine W. E., Elliker P. R. Involvement of phosphoenolpyruvate in lactose utilization by group N streptococci. J Bacteriol. 1969 Aug;99(2):603–610. doi: 10.1128/jb.99.2.603-610.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L., Miller A., 3rd, Sandine W. E., Elliker P. R. Mechanisms of lactose utilization by lactic acid streptococci: enzymatic and genetic analyses. J Bacteriol. 1970 Jun;102(3):804–809. doi: 10.1128/jb.102.3.804-809.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse M. L., Hill K. L., Egan J. B., Hengstenberg W. Metabolism of lactose by Staphylococcus aureus and its genetic basis. J Bacteriol. 1968 Jun;95(6):2270–2274. doi: 10.1128/jb.95.6.2270-2274.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve E. C., Braithwaite J. A. Lac-plus plasmids are responsible for the strong lactose-positive phenotype found in many strains of Klebsiella species. Genet Res. 1973 Dec;22(3):329–333. doi: 10.1017/s0016672300013124. [DOI] [PubMed] [Google Scholar]
- Warren R. A. Lactose-utilizing mutants of lac deletion strains of Escherichia coli. Can J Microbiol. 1972 Sep;18(9):1439–1444. doi: 10.1139/m72-221. [DOI] [PubMed] [Google Scholar]




