Abstract
Recently, mutations in the Met tyrosine kinase receptor have been identified in both hereditary and sporadic forms of papillary renal carcinoma. We have introduced the corresponding mutations into the met cDNA and examined the effect of each mutation in biochemical and biological assays. We find that the Met mutants exhibit increased levels of tyrosine phosphorylation and enhanced kinase activity toward an exogenous substrate when compared with wild-type Met. Moreover, NIH 3T3 cells expressing mutant Met molecules form foci in vitro and are tumorigenic in nude mice. Enzymatic and biological differences were evident among the various mutants examined, and the somatic mutations were generally more active than those of germ-line origin. A strong correlation between the enzymatic and biological activity of the mutants was observed, indicating that tumorigenesis by Met is quantitatively related to its level of activation. These results demonstrate that the Met mutants originally identified in human papillary renal carcinoma are oncogenic and thus are likely to play a determinant role in this disease, and these results raise the possibility that activating Met mutations also may contribute to other human malignancies.
The Met tyrosine kinase is a high-affinity receptor for hepatocyte growth factor/scatter factor (HGF/SF) (1, 2). Both Met and HGF/SF are expressed in numerous tissues, although their expression is confined predominantly to cells of epithelial and mesenchymal origin, respectively (3, 4). Signaling via this receptor-ligand pair has been shown to affect a wide range of biological activities, including angiogenesis (5, 6), cellular motility (3), growth (7–9), invasion (10–12), and morphogenic differentiation (13–15). Met-HGF/SF signaling also has been shown to be essential for normal murine embryological development (16–18) and is believed to play a role in tissue regeneration (19), wound healing (20), and the development of various organs (16, 17, 21–25).
Whereas Met-HGF/SF signaling clearly mediates a variety of normal cellular processes, this receptor-ligand pair also has been implicated in the generation and spread of tumors (reviewed in ref. 26). Met originally was isolated as the product of a human oncogene, tpr-met, which encodes an altered Met protein possessing constitutive kinase activity and transforming ability (27, 28). The coexpression of unaltered Met and HGF/SF molecules in the same cell, which generates an autocrine stimulatory loop, is also oncogenic (12, 29–32). Although the inappropriate expression of these molecules has been documented in a wide variety of human tumors (reviewed in ref. 26), conclusive evidence that Met and/or HGF/SF play a role in human malignancy has been lacking.
Recently, however, evidence implicating Met in the development of human papillary renal carcinoma was obtained (33). Papillary renal carcinomas, which comprise approximately 14% of all renal cell neoplasms, are recognized histologically by the presence of vascularized connective tissue stalks surrounded by neoplastic cells (34). Both hereditary and sporadic forms of the disease have been identified. A gene associated with the hereditary form of the disease was mapped to a region encompassing the met locus, and sequencing revealed germ-line Met mutations in affected individuals (33). Somatic Met mutations also were found in some sporadic cases of the disease. All of the mutations identified were missense mutations that localized to the tyrosine kinase domain of the Met receptor. To investigate the possibility that these tumor-associated mutations caused constitutive activation of the Met receptor, we introduced the mutations into the met cDNA and examined their activity in biochemical and biological assays.
MATERIALS AND METHODS
Cell Lines.
NIH 3T3 cells (CRL 1658) were obtained from the American Type Culture Collection and cultured in DMEM (Life Technologies)/10% calf serum (CS) (Life Technologies). These cells produce only two scatter units/ml of HGF/SF, which is significantly lower than other NIH 3T3 sublines we have analyzed (data not shown).
Constructs.
The pMB1 expression vector (29), which uses the Moloney murine sarcoma virus long terminal repeat promoter, was used. The wild-type Met expression vector (called pMB11) contains the murine Met cDNA in pMB1 (29). To construct the Met mutants, the QuikChange site-directed mutagenesis kit (Stratagene) was used according to the manufacturer’s instructions with pMB11 as the template. Mutations were verified by sequencing both strands of DNA in the region of interest.
Transfections and Focus-Formation Assay.
Transfections were performed using Lipofectamine according to the manufacturer’s protocol (Life Technologies). Cells (1.5 × 105) in 35-mm plastic dishes were transfected with 2 μg of DNA containing 1.77 μg of the plasmid of interest and .23 μg of a plasmid (pSV2neo; ref. 35) conferring resistance to G418 (Life Technologies). Three days after transfection, cells were split into one 140-mm dish containing DMEM/5% CS and one containing DMEM/10% CS supplemented with 800 μg/ml G418 (Life Technologies). The cultures were fed every 3–4 days. After 2 weeks, the cells cultured in DMEM/5% CS were stained with .2% crystal violet in 70% ethanol, and foci were counted and photographed. The cells cultured in DMEM/10% CS supplemented with G418 were used to assess transfection efficiency and were grown as pools of cells consisting of at least 100 colonies and used for expression, phosphorylation, and tumorigenesis experiments. Because all of the constructs generated comparable numbers of G418-resistant colonies, differences in focus-forming ability are not due to differences in transfection efficiencies between the constructs.
Western Blotting.
Western analysis was performed essentially as described (12) under reducing conditions using the following primary antibodies: anti-Met (SP260; Santa Cruz Biotechnology), anti-phosphotyrosine (see Fig. 1A; Second from Top) (4G10; a gift of Deborah Morrison, Advanced Bioscience Laboratories, National Cancer Institute-Frederick Cancer Research and Development Center, Frederick, MD), and anti-phosphotyrosine (see Fig. 1A; Third from Top) (PY20; Transduction Laboratories, Lexington, KY).
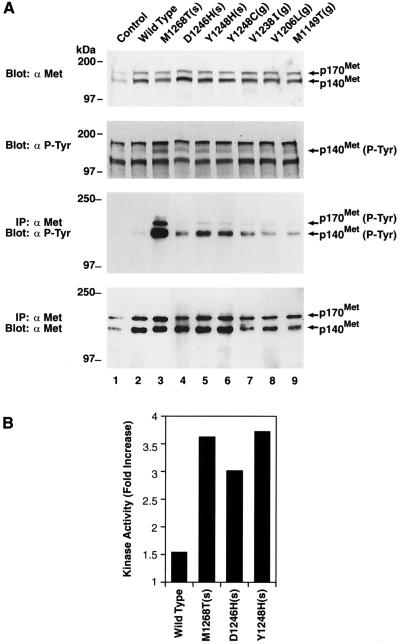
Figure 1.
Expression, autophosphorylation, and exogenous kinase activity of wild-type and mutant Met in NIH 3T3 cells. Samples labeled control and wild type are from cells stably transfected with empty vector or vector expressing wild-type Met, respectively. All other samples are from cells stably transfected with vectors expressing the indicated Met mutant. Cells were cultured in DMEM/10% CS before harvest. (A, Top) Fifty micrograms of cell lysate/sample were resolved on a 7.5% gel and examined by Western analysis using anti-Met antibody. (A, Second from Top) The filter was stripped and reprobed with anti-phosphotyrosine antibody. (A, Third from Top) Four hundred micrograms of cell lysate/sample were immunoprecipitated with anti-Met antibody, resolved on an 8% gel, and analyzed by Western analysis using anti-phosphotyrosine antibody. (A, Bottom) The filter was stripped and reprobed with anti-Met antibody. Molecular mass markers are indicated on the left. (B) Two hundred micrograms of cell lysate/samples were immunoprecipitated with anti-Met antibody and assessed for kinase activity toward an exogenous substrate (gastrin) using a tyrosine kinase assay kit. Results are reported as fold increase relative to cells transfected with empty vector. Samples were performed in triplicate, and SDs were ≤ 5% of the mean.
Immunoprecipitation.
Monolayers of stably transfected cells were washed 2× with ice-cold PBS, lysed in ice-cold buffer consisting of 20 mM Tris (pH 8.0), 137 mM NaCl, 10% glycerol, .1% SDS, .5% Nonidet P-40, 100 mM sodium fluoride, 200 uM sodium orthovanadate, 1 mM EGTA, .2 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 3 μg/ml aprotinin, and centrifuged (30 min, 4°C, 16,000 × g). After quantitation, 400 μg of each lysate was precleared with protein A Sepharose and then incubated with anti-Met antibody (SP260; Santa Cruz Biotechnology) and protein A Sepharose for 3 hr at 4°C with rotation. The samples then were washed 3× with ice-cold buffer consisting of 20 mM Tris (pH 7.4), 100 mM NaCl, .5% Nonidet P-40, and 1 mM EDTA, and 2× with ice-cold PBS. SDS gel-loading buffer (containing reducing agent) then was added to each sample. After boiling (5 min) and centrifuging (5 min, 16,000 × g), the resulting supernatants were resolved by SDS/PAGE and examined by Western analysis.
Nonradioactive Tyrosine Kinase Assay.
Cell lysates were prepared and immunoprecipitated using anti-Met SP260 (Santa Cruz Biotechnology) antibody as described (36). Immunoprecipitates were assessed for tyrosine kinase activity toward an exogenous substrate using a nonradioactive tyrosine kinase assay kit according to the manufacturer’s instructions (Boehringer Mannheim). Standard curves were constructed to verify the linear range of the assay, and all samples fell within the linear range.
In Vivo Tumorigenicity Assay and Generation of Tumor Explants.
Pools of G418-resistant NIH 3T3 cells expressing the indicated Met protein were generated as described above and were shown to express ≈equal levels of exogenous Met before use. The cells were prepared as described (30), and 5 × 105 cells were inoculated subcutaneously into ≈4-week-old female athymic nude mice. To develop explants, 200- to 400-mm2 tumors (see Table 1) were minced and cultured for ≈2 weeks in DMEM/10% CS supplemented with 800 μg/ml G418. Two independently derived explants generated from each construct (one of which is illustrated in Fig. 3) exhibited similar levels of Met expression and were phenotypically similar.
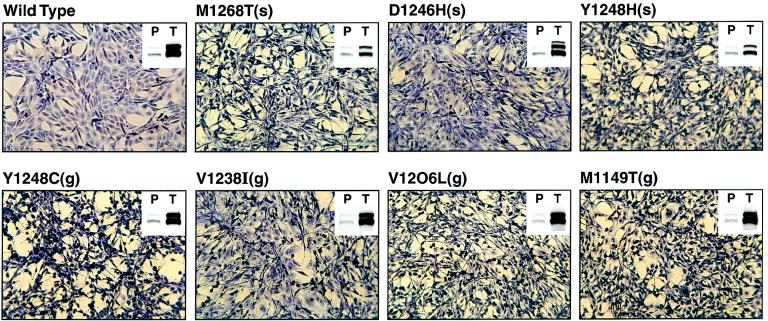
Figure 3.
Morphology of and Met expression in explants derived from tumors induced by NIH 3T3 cells expressing wild-type or mutant Met. Explants prepared from tumors induced by cells expressing the indicated construct were photographed at a magnification of ×100. (Insets) Fifty micrograms of cell lysate/sample were resolved on a 7.5% gel and examined by Western analysis using anti-Met antibody. P, samples from a pool of cells, transfected with the indicated construct, that were used as the inoculum for tumor induction. T, samples from explants prepared from tumors induced by cells transfected with the indicated construct.
RESULTS
The Met mutants are labeled according to amino acid change, location, and germ-line (g) vs. somatic (s) status; thus mutant M1268T(s) corresponds to a methionine to threonine change at amino acid number 1,268 of the Met protein that had been identified as a somatic mutation (33). Upon transfection into NIH 3T3 cells followed by G418 selection and Western analysis with anti-Met antibody (Fig. 1A, Top), we found that cells transfected with wild-type (lane 2) and mutant (lanes 3–9) Met constructs express comparable levels of exogenous Met protein that is significantly greater than the small quantity of endogenous Met expressed by control cells transfected with empty vector (lane 1). The 170-kDa product detected corresponds to the intracellular single-chain Met precursor, whereas the 140-kDa product corresponds to the β-chain of the mature, cell surface-associated 190-kDa disulfide-linked Met heterodimer (37, 38).
Phosphorylation of Met on tyrosine residues has been shown to activate its intrinsic kinase activity (39, 40) and therefore is an indicator of enzymatic activity. To examine the phosphorylation status of Met, the filter presented in Fig. 1A (Top) was stripped and reprobed with anti-phosphotyrosine antibody (Fig. 1A; Second from Top). A product corresponding in size to p140Met was observed in lysates from cells expressing the mutant Met proteins, but not from cells expressing wild-type Met or control cells transfected with empty vector. Thus, mutant Met proteins appear to be in a more activated state than wild-type Met. To confirm and extend this finding, lysates from cells transfected with the mutant Met constructs first were immunoprecipitated with anti-Met antibody and then examined by Western analysis with anti-phosphotyrosine antibody (Fig. 1A; Third from Top). These results demonstrate that the mutant Met proteins are indeed phosphorylated to a greater degree than wild-type Met. A reprobing of the filter with anti-Met antibody demonstrates that the phosphorylation of the mutant Met proteins is not due to their increased expression relative to wild-type Met (Fig. 1A, Bottom). It is clear from this data that whereas all of the mutants induce more autophosphorylation than does wild-type Met, one mutant [i.e., M1268T(s)] is very strongly activating, whereas others are only moderately or weakly activating (see Table 1). We also found that anti-Met immunoprecipitates obtained from cells expressing mutant Met proteins possess greater tyrosine kinase activity toward an exogenous substrate than do immunoprecipitates obtained from cells expressing wild-type Met (Fig. 1B).
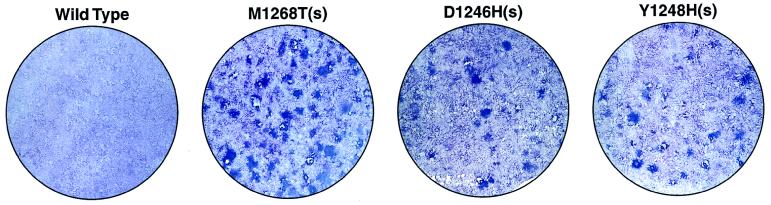
We noted that cells stably transfected with some of the Met mutants [i.e., M1268T(s), D1246H(s), Y1248H(s), and Y1248C(g)] were phenotypically transformed, whereas cells stably transfected with wild-type Met and some of the other Met mutants [V1238I(g), V1206L(g), and M1149T(g)] were phenotypically normal (data not shown). To quantitate the in vitro transforming ability of the mutants we performed a focus-formation assay. The subline of NIH 3T3 cells used for this assay produces only a very small quantity of HGF/SF (see Materials and Methods), such that wild-type Met does not generate any foci in these cells via autocrine stimulation (Fig. 2 and Table 1). In contrast, five of the eight Met mutants examined induce significant focus formation (Fig. 2 and Table 1). We observed a good correlation between enzymatic activity (see above) and focus-forming activity, with the most enzymatically active mutant [M1268T(s)] exhibiting the strongest focus-forming activity (>300 foci/μg). In addition, four of the enzymatically moderate mutants [D1246N(g), D1246H(s), Y1248H(s), and Y1248C(g)] were competent for focus formation (115–156 foci/μg), whereas three enzymatically moderate to weak mutants [V1238I(g), V1206L(g), and M1149T(g)] were negative for focus formation.
Figure 2.
Focus induction by wild-type and mutant Met in NIH 3T3 cells. Cells transfected with the indicated constructs were assessed for focus-forming ability. A representative field of view is shown for each sample. See Table 1 for additional information on focus-forming activity.
We also tested NIH 3T3 cells expressing each of the Met proteins for tumorigenicity in athymic nude mice. We found that cells expressing each of the mutant Met proteins form tumors more readily than do cells expressing wild-type Met (Table 1). Histologically, the tumors were poorly differentiated; locally invasive sarcomas and DNAs extracted from the tumors were shown to contain the expected exogenous mutant Met sequence (not shown). As with the focus-formation assay, a strong correlation between enzymatic activity and tumorigenesis was evident, with the most enzymatically active mutant [M1268T(s)] also proving to be the most tumorigenic. In addition, all of the enzymatically moderate to weak mutants, even those that lacked focus-forming activity, were more tumorigenic than wild-type Met. The greater sensitivity of the nude mouse assay relative to the focus-formation assay for detecting transforming activity has been described (41).
Cultured explants derived from tumors induced by cells expressing wild-type Met, which form tumors after a longer latency period than the Met mutants (Table 1), appear phenotypically normal, whereas explants derived from tumors induced by cells expressing mutant Met are phenotypically transformed (Fig. 3). Moreover, explants derived from tumors induced by the various mutant Met proteins are themselves phenotypically heterogeneous. An examination of Met expression in the explants reveals that, without exception, the explanted cells express significantly higher levels of Met than do the pools of cells comprising the respective tumor inoculum (Fig. 3; Insets). The selection of cells expressing high levels of Met during tumor formation supports a causative role for Met in tumor induction and has been previously observed (29, 42). Interestingly, the level of Met expressed in explants derived from tumors induced by the most enzymatically and biologically active mutant [M1268T(s)] was lower than that exhibited by explants derived from tumors induced by the other Met mutants or by wild-type Met. Mutant Y1248H(s), which exhibits the second strongest biological activity of all the mutants, also displayed a reduced explant-associated Met expression relative to the other mutants. This finding may indicate that tumor induction by the most active Met mutants requires less protein product than does tumor induction by the less active Met mutants and by wild-type met. However, the possibility that the decreased Met expression exhibited by the most active Met mutants is due to a higher turnover rate cannot be discounted.
DISCUSSION
Our findings demonstrate that the Met mutations that previously had been identified in both hereditary and sporadic cases of human papillary renal carcinoma stimulate the enzymatic and biological activity of this tyrosine kinase receptor. As a group, the somatic mutations that had been identified in sporadic cases of this disease are comparatively more activating than the germ-line mutations associated with hereditary cases of the disease (Table 1). It is possible that highly activating Met mutations may be incompatible with life if introduced into the germ-line.
Why is papillary renal carcinoma the predominant form of malignancy in individuals harboring the activating germ-line Met mutations analyzed in the present study? Met is highly expressed in the kidney (43, 44), and Met-HGF/SF signaling has been shown to mediate both mitogenic (45, 46) and morphogenic (13) programs in cultured kidney cells. Thus, Met expression level and kidney-associated biological consequences of Met signaling may be important contributing factors. However, Met also is highly expressed (43, 44) and capable of inducing growth (9) and morphogenic alterations (14) in a number of other tissues. Interestingly, malignancies other than papillary renal carcinoma (i.e., carcinomas of the stomach, rectum, lung, pancreas, breast, and bile duct), all of which occur in Met-expressing tissues, have been identified in some of the individuals with activating germ-line Met mutations (33, 47). Thus, although papillary renal carcinoma is the predominant form of malignancy in individuals harboring activating germ-line Met mutations, mutant Met also may play a role in the development of other cancers in these individuals. Furthermore, the possibility that activating somatic Met mutations contribute to the formation of tumors other than papillary renal carcinoma deserves consideration.
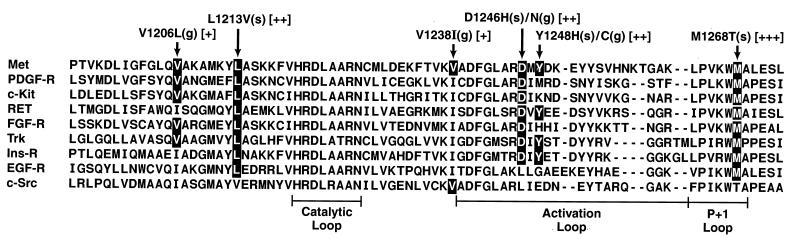
We note that based on the structural homology among protein kinases (48, 49), some of the Met mutations described in the present analysis are likely to be activating when introduced into other kinase molecules. In fact, the Ret (50) and Kit (51, 52) receptor tyrosine kinases have been shown to be activated by amino acid changes at methionine and aspartic acid residues corresponding to the positions of Met mutants M1268 and D1246, respectively (Fig. 4). Because some of the residues corresponding to the positions of the other Met mutants are conserved in a number of other tyrosine kinase receptors (Fig. 4), the identification of novel activating Met mutations raises the possibility that comparable mutations in other protein kinases may play a role in human malignancy.
Figure 4.
An alignment of a portion of the COOH-terminal lobe of a cytoplasmic tyrosine kinase (c-src) and representative receptor tyrosine kinases (all other samples) (49, 53). The locations and relative activities of seven of the Met mutations analyzed are shown above affected residues. Conserved amino acids are shown in black. Residues, which, when mutated, activate the Kit and Ret receptors, are shown in broken black boxes. See text for additional details. See note added in proof.
The mechanisms by which the various mutations activate the transforming potential of the Met receptor remain to be elucidated. Seven of the eight activating Met mutations fall within the COOH-terminal lobe of the kinase domain (see Fig. 4). Among other things, this region of the molecule is believed to act as an intramolecular substrate, which, in the absence of ligand, functions to inhibit enzymatic activity by blocking the active site (53). It is possible that some of the mutations stimulate the kinase activity of Met by altering the structure of the intramolecular substrate such that it is constitutively disengaged from the active site.
Deregulated kinase activity may not, however, be the sole event necessary for activating the transforming potential of the Met receptor. Rather, a qualitative shift in substrate specificity also may play an important role. Such a scenario has been suggested for the aforementioned Ret (54) and Kit (55) mutations corresponding to the positions of Met mutants M1268 and D1246, respectively. In both cases, the mutations change a residue conserved in receptor tyrosine kinases to one typical of nonreceptor tyrosine kinases and may alter receptor conformations such that they now interact with SH2-domain-containing proteins, which normally preferentially interact with nonreceptor tyrosine kinases. It is possible that a similar mechanism is involved in activating the transforming potential of the Met mutant receptors.
Table 1.
Activity of Met mutants
| Met construct* | Met phos-phorylation† | Focus formation‡ #foci/μg DNA | Tumor formation§
|
|
|---|---|---|---|---|
| # mice with tumors/# mice injected | Mean tumor size, mm2 | |||
| Wild type | − | 0 | 0/14¶ | 0¶ |
| M1268T(s) | +++ | >300 | 8/8 | 216 ± 77 |
| Y1248H(s) | ++ | 156 ± 16 | 8/8 | 100 ± 40 |
| D1246H(s) | ++ | 119 ± 16 | 8/8 | 60 ± 52 |
| D1246N(g) | ++ | 147 ± 5 | 9/9 | 50 ± 25 |
| Y1248C(g) | ++ | 115 ± 11 | 7/8 | 77 ± 89 |
| V1238l(g) | ++ | 0 | 5/8 | 13 ± 15 |
| V1206L(g) | + | 0 | 6/6 | 50 ± 32 |
| M1149T(g) | + | 0 | 4/8 | 46 ± 56 |
See legend to Fig. 1.
Data from Fig. 1A (and data not shown).
NIH 3T3 cells transfected with the indicated constructs were scored for focus formation after 2 weeks. Results represent the mean of two independent experiments. Also see Fig. 2.
NIH 3T3 cells transfected with the indicated constructs were inoculated subcutaneously into nude mice, and tumors were measured after 2 weeks. Results represent the mean of two independent experiments.
Cells transfected with wild-type Met do eventually form tumors, probably due to the generation of an autocrine Met-HGF/SF stimulatory loop, as we previously described (29). However, tumors derived from cells expressing wild-type Met do not appear until ∼4 weeks after inoculation, by which time most mice inoculated with cells expressing mutant Met molecules have had to be sacrificed due to tumor burden.
Acknowledgments
We thank Linda Miller, Marilyn Powers, Leo Lee, Terry Sweeney, and Oscar Smith for technical assistance; Richard Frederickson for performing the artwork and photography; and Ave Cline for typing the manuscript. Research was sponsored in part by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories.
ABBREVIATIONS
- HGF/SF
hepatocyte growth factor/ scatter factor
- CS
calf serum
Note Added in Proof
We recently have found that Met possessing the previously described L1213V(S) mutation (33) is capable of focus induction in NIH 3T3 Cells.
References
- 1.Bottaro D P, Rubin J S, Faletto D L, Chan A M, Kmiecik T E, Vande Woude G F, Aaronson S A. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 2.Naldini L, Weidner K M, Vigna E, Gaudino G, Bardelli A, Ponzetto C, Narsimhan R P, Hartmann G, Zarnegar R, Michalopoulos G K, Birchmeier W, Comoglio P M. EMBO J. 1991;10:2867–2878. doi: 10.1002/j.1460-2075.1991.tb07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoker M, Gherardi E, Perryman M, Gray J. Nature (London) 1987;327:239–242. doi: 10.1038/327239a0. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenberg E, Meyer D, Weidner K M, Birchmeier C. J Cell Biol. 1993;123:223–235. doi: 10.1083/jcb.123.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussolino F, Di Renzo M F, Ziche M, Bocchietto E, Olivero M, Naldini L, Gaudino G, Tamagnone L, Coffer A, Comoglio P M. J Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant D S, Kleinman H K, Goldberg I D, Bhargava M M, Nickloff B J, Kinsella J L, Polverini P, Rosen E M. Proc Natl Acad Sci USA. 1993;90:1937–1941. doi: 10.1073/pnas.90.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashio K, Shima N. In: Hepatocyte Growth Factor-Scatter Factor and the c-Met Receptor. Goldberg I D, Rosen E M, editors; Goldberg I D, Rosen E M, editors. Vol. 65. Basel: Birkhauser; 1993. pp. 351–368. [Google Scholar]
- 8.Nakamura T, Teramoto H, Ichihara A. Proc Natl Acad Sci USA. 1986;83:6489–6493. doi: 10.1073/pnas.83.17.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin J S, Chan A M, Bottaro D P, Burgess W H, Taylor W G, Cech A C, Hirschfield D W, Wong J, Miki T, Finch P W, Aaronson S A. Proc Natl Acad Sci USA. 1991;88:415–419. doi: 10.1073/pnas.88.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidner K M, Behrens J, Vandekerckhove J, Birchmeier W. J Cell Biol. 1990;111:2097–2108. doi: 10.1083/jcb.111.5.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rong S, Segal S, Anver M, Resau J H, Vande Woude G F. Proc Natl Acad Sci USA. 1994;91:4731–4735. doi: 10.1073/pnas.91.11.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffers M, Rong S, Vande Woude G F. Mol Cell Biol. 1996;16:1115–1125. doi: 10.1128/mcb.16.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montesano R, Matsumoto K, Nakamura T, Orci L. Cell. 1991;67:901–908. doi: 10.1016/0092-8674(91)90363-4. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann V, Foroutan H, Sachs M, Weidner K M, Birchmeier W. J Cell Biol. 1995;131:1573–1586. doi: 10.1083/jcb.131.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsarfaty I, Resau J H, Rulong S, Keydar I, Faletto D L, Vande Woude G F. Science. 1992;257:1258–1261. doi: 10.1126/science.1387731. [DOI] [PubMed] [Google Scholar]
- 16.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Nature (London) 1995;375:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Nature (London) 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 18.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Nature (London) 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto K, Nakamura T. In: Hepatocyte Growth Factor-Scatter Factor and the Met Receptor. Goldberg I D, Rosen E M, editors; Goldberg I D, Rosen E M, editors. Vol. 65. Basel: Birkhauser; 1993. pp. 225–250. [Google Scholar]
- 20.Nusrat A, Parkos C A, Bacarra A E, Godowski P J, Delp-Archer C, Rosen E M, Madara J L. J Clin Invest. 1994;93:2056–2065. doi: 10.1172/JCI117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos O F P, Barros E J G, Yang X-M, Matsumoto K, Nakamura T, Park M, Nigam S K. Dev Biol. 1994;163:525–529. doi: 10.1006/dbio.1994.1169. [DOI] [PubMed] [Google Scholar]
- 22.Woolf A S, Kolatsi-Joannou M, Hardman P, Andermarcher E, Moorby C, Fine L G, Jat P S, Noble M D, Gherardi E. J Cell Biol. 1995;128:171–184. doi: 10.1083/jcb.128.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Spitzer E, Meyer D, Sachs M, Niemann C, Hartmann G, Weidner K M, Birchmeier C, Birchmeier W. J Cell Biol. 1995;131:1–12. doi: 10.1083/jcb.131.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soriano J V, Pepper M S, Nakamura T, Orci L, Montesano R. J Cell Sci. 1995;108:413–430. doi: 10.1242/jcs.108.2.413. [DOI] [PubMed] [Google Scholar]
- 25.Streit A, Stern C D, Thery C, Ireland G W, Aparicio S, Sharpe M J, Gherardi E. Development (Cambridge, UK) 1995;121:813–824. doi: 10.1242/dev.121.3.813. [DOI] [PubMed] [Google Scholar]
- 26.Jeffers M, Rong S, Vande Woude G F. J Mol Med. 1996;74:505–513. doi: 10.1007/BF00204976. [DOI] [PubMed] [Google Scholar]
- 27.Cooper C S, Park M, Blair D G, Tainsky M A, Huebner K, Croce C M, Vande Woude G F. Nature (London) 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 28.Park M, Dean M, Kaul K, Braun M J, Gonda M A, Vande Woude G F. Proc Natl Acad Sci USA. 1987;84:6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rong S, Bodescot M, Blair D, Dunn J, Nakamura T, Mizuno K, Park M, Chan A, Aaronson S, Vande Woude G F. Mol Cell Biol. 1992;12:5152–5158. doi: 10.1128/mcb.12.11.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffers M, Rong S, Anver M, Vande Woude G F. Oncogene. 1996;13:853–861. [PubMed] [Google Scholar]
- 31.Kanda H, Tajima H, Lee G-H, Nomura K, Ohtake K, Matsumoto K, Nakamura T, Kitagawa T. Oncogene. 1993;8:3047–3053. [PubMed] [Google Scholar]
- 32.Bellusci S, Moens G, Gaudino G, Comoglio P, Nakamura T, Thiery J-P, Jouanneau J. Oncogene. 1994;9:1091–1099. [PubMed] [Google Scholar]
- 33.Schmidt L, Duh F-M, Chen F, Kishida T, Glenn G, et al. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 34.Weiss L M, Gelb A B, Medeiros L J. Anat Pathol. 1994;103:624–635. doi: 10.1093/ajcp/103.5.624. [DOI] [PubMed] [Google Scholar]
- 35.Southern P J, Berg P. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 36.Webb C P, Lane K, Dawson A P, Vande Woude G F, Warn R M. J Cell Sci. 1996;109:2371–2381. doi: 10.1242/jcs.109.9.2371. [DOI] [PubMed] [Google Scholar]
- 37.Faletto D L, Tsarfaty I, Kmiecik T E, Gonzatti M, Suzuki T, Vande Woude G F. Oncogene. 1992;7:1149–1157. [PubMed] [Google Scholar]
- 38.Giordano S, Di Renzo M F, D R, Narsimhan R P, Cooper C S, Rosa C, Comoglio P M. Oncogene. 1989;4:1383–138. [PubMed] [Google Scholar]
- 39.Naldini L, Vigna E, Ferracini R, Longati P, Gandino L, Prat M, Comoglio P M. Mol Cell Biol. 1991;11:1793–1803. doi: 10.1128/mcb.11.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodrigues G A, Park M. Oncogene. 1994;9:2019–2027. [PubMed] [Google Scholar]
- 41.Blair D G, Cooper C S, Oskarsson M K, Eader L A, Vande Woude G F. Science. 1982;218:1122–1125. doi: 10.1126/science.6293052. [DOI] [PubMed] [Google Scholar]
- 42.Rong S, Oskarsson M, Faletto D, Tsarfaty I, Resau J H, Nakamura T, Rosen E, Hopkins R F R, Vande Woude G F. Cell Growth Differ. 1993;4:563–569. [PubMed] [Google Scholar]
- 43.Iyer A, Kmiecik T E, Park M, Daar I, Blair D, Dunn K J, Sutrave P, Ihle J N, Bodescot M, Vande Woude G F. Cell Growth Diff. 1990;1:87–95. [PubMed] [Google Scholar]
- 44.Di Renzo M F, Narsimhan R P, Olivero M, Bretti S, Giordano S, Medico E, Gaglia P, Zara P, Comoglio P M. Oncogene. 1991;6:1997–2003. [PubMed] [Google Scholar]
- 45.Igawa T, Kanda S, Kanetake H, Saitoh Y, Ichihara A, Tomita Y, Nakamura T. Biochem Biophys Res Commun. 1991;174:831–838. doi: 10.1016/0006-291x(91)91493-v. [DOI] [PubMed] [Google Scholar]
- 46.Kan M, Zhang G, Zarnegar R, Michalopoulos G, Myoken Y, McKeehan W L, Stevens J I. Biochem Biophys Res Commun. 1991;174:331–337. doi: 10.1016/0006-291x(91)90524-b. [DOI] [PubMed] [Google Scholar]
- 47.Zbar B, Tory K, Merino M, Schmidt L, Glenn G, Choyke P, Walther M M, Lerman M, Linehan W M. J Urol. 1994;151:561–566. doi: 10.1016/s0022-5347(17)35015-2. [DOI] [PubMed] [Google Scholar]
- 48.Hanks S K, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 49.Hanks S K. Curr Opin Struct Biol. 1991;1:369–383. [Google Scholar]
- 50.Santoro M, Carlomagno F, Romano A, Bottaro D P, Dathan N A, Grieco M, Fusco A, Vecchio G, Matoskova B, Kraus M H, Di Fiore P. Science. 1995;267:381–383. doi: 10.1126/science.7824936. [DOI] [PubMed] [Google Scholar]
- 51.Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfield J H, Ashman L K, Kanayama Y, Matsuzawa Y, Kitamura Y, Kanakura Y. J Clin Invest. 1993;92:1736–1744. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piao X, Bernstein A. Blood. 1996;87:3117–3123. [PubMed] [Google Scholar]
- 53.Hubbard S R, Wei L, Ellis L, Hendrickson W A. Nature (London) 1994;372:22–29. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- 54.Songyang Z, Carraway K L, Eck M J, Harrison S C, Feldman R A, Mohammadi M, Schlessinger J, Hubbard S R. Nature (London) 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 55.Piao X, Paulson R, van der Geer P, Pawson T, Bernstein A. Proc Natl Acad Sci USA. 1996;93:14665–14669. doi: 10.1073/pnas.93.25.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]