Abstract
Zebra finches have been widely used to study neurobiology underlying vocal development. Because only male zebra finches learn song, efficient developmental use of these animals requires early determination of sex at ages that precede maturation of secondary sex characteristics. We have developed a sex determination method that combines a forensics method of genomic DNA isolation (from very small blood samples) with PCR amplification from Z and W sex chromosomes (males are ZZ, females ZW). This combination results in a minimally-invasive yet highly reliable and convenient genotyping method.
Introduction
Because juvenile zebra finches learn to sing in distinct stages, they have become an important model for studying the neurobiology underlying vocal development. We have used these animals to study effects of CNS-active drugs during late-postnatal development (Soderstrom and Johnson, 2003; Soderstrom and Johnson, 2000; Soderstrom and Johnson, 2001; Soderstrom and Tian, 2006, 2004; Soderstrom et al., 2004). As our experiments employ young birds prior to maturation of secondary sex characteristics, and only males of the species undergo sensory-motor vocal learning, it became important for us to develop a reliable method for determining the sex of zebra finch fledglings that is amenable to application of minimally-invasive blood sampling techniques and high-throughput use. We have accomplished these goals by adapting forensics methods for isolation of genomic DNA from fabric-absorbed blood droplets and combining them with sexing methods based on PCR amplification from zebra finch Z and W sex chromosomes.
A good general method for determining the sex of birds has previously been described (Griffiths et al., 1998). This method relies on PCR amplification using a single pair of degenerate primers (designated P2 and P8) targeting isoforms of the CHD (chromohelicase-DNA-binding) genes that reside on avian sex chromosomes (females are ZW, males ZZ). Due to differing intron lengths within the CHD coding sequence, avian genomic DNA derived from ZW females typically results in PCR amplification of two distinctly-sized fragments, whereas ZZ male-derived DNA results in a single product. The degenerate nature of the PCR primers designed by Griffiths, et al is advantageous in that it allows use of a single pair of primers to sex a wide variety of avian species.
The size of Z- and W-derived P2- and P8-primed CHD gene amplification products differ across avian species. In some avian species CHD sequences are missing entirely (de Kloet and de Kloet, 2003). In the case of zebra finches, CHD is present and the difference between male and female P2- and P8-amplified isoforms is 36 bp, which is easily resolved on standard 2% agarose gels. Unfortunately, for reasons that likely involve the degenerate nature of the P2 and P8 primers employed, use of this single primer approach with zebra finch genomic DNA as a template, even optimal reaction conditions result in preferential amplification from the shared Z chromosome (e.g. see (Griffiths et al., 1998), Fig 1). Frequently, amplification from the female-specific W chromosome is undetectable, resulting in erroneous sex assignment.
Figure 1.
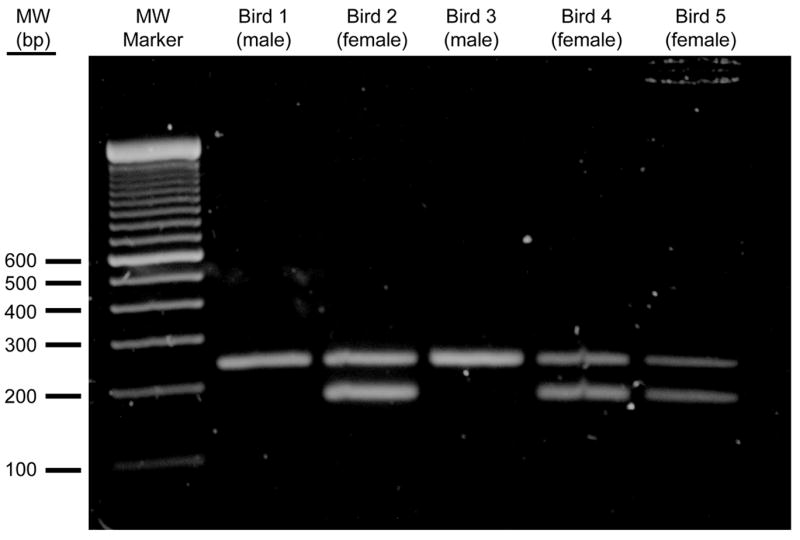
Other PCR-based methods for zebra finch sex assignment from amplification of large-scale isolation of genomic DNA have been reported (Runciman et al., 1999). We have developed a similar method of direct amplification of male- and female-derived genomic DNA that results in unequivocal sex determination by combining two sets of zebra finch sequence specific PCR primers in single reactions: one pair specifically targets the female W chromosome, while the other pair targets the shared Z chromosome. Thus, amplification of male genomic DNA (ZZ) results in a single product of 242 bp, while that of female DNA (ZW) results in amplification from both Z and W chromosomes and production of two products of 242 bp and 179 bp, respectively. Problems related to preferential amplification of one of the two female sex chromosomes have not been experienced (e.g. Fig 1). The 63 bp difference between these products make them easy to resolve on standard 1.2% agarose gels.
The combination of DNA isolation from trace blood samples obtained through minimally-invasive procedures, and a PCR method that has been optimized for amplification from template prepared in this manner, makes the integrated approach described herein distinctly applicable to high-throughput sex determination required by large breeding aviaries.
Methods
Enzymes and Reagents
Oligonucleotide PCR primers were synthesized by Invitrogen. Chelex 100 resin was purchased from Bio-Rad (Cat # 142–1253). All other supplies, enzymes and reagents were purchased from either Fisher or Sigma.
Isolation of genomic DNA
We adapted the forensic methods described by (Walsh et al., 1991) to isolate and purify zebra finch genomic DNA for use as a template for PCR. This method is designed to reliably extract DNA from very small amounts of blood absorbed by fabric, and thus works well for processing small blood samples collected from zebra finches following minimally invasive procedures.
Zebra finches are captured and firmly held with the thumb and third finger securing splayed legs. Next, a leg is wiped with a 70% ETOH-soaked pad and the region just distal from the knee is pricked with a fine 30 gauge needle. The resulting blood droplet is absorbed with a small chad (approximately 3 mm2) of sterile 3MM chromatography paper held in forceps (see Fig 2). In addition to collecting blood, the paper tends to stop bleeding at the lance site. The blood-containing paper is placed into a sterile 1.5 ml centrifuge tube for further processing and the bird is released. The entire collection procedure can be completed within a matter of seconds.
Figure 2.

Zebra finch blood sample collection. A young zebra finch is held gently, but firmly in the hand. The left leg is held between thumb and forefinger, swabbed with 70% ETOH, and a small prick is made with a sterile hypodermic needle just distal of the elbow joint. The resulting droplet of blood is collected with an autoclaved chad of chromatography paper held by forceps, as shown. Bleeding is usually stopped following absorption. The bird is then released and the sample-containing paper is placed in a sterile 1.5 ml centrifuge tube for isolation of genomic DNA as described in the text. With experience the entire sampling procedure can be completed in a matter of seconds.
To each blood sample-containing tube, 1 ml of distilled water is added and mixed gently by inversion. These samples are allowed to sit at room temperature with occasional mixing (to suspend blood cells from the paper matrix) for 30 min. Following suspension, blood cells are collected by centrifugation at 15,000 × g for 3 min. The supernatant is carefully removed and discarded leaving about 20 mcl (containing collected blood cells) and the paper chad undisturbed at the bottom of the tube. 200 mcl of a 5% suspension (wt/vol, in sterile distilled water) of Chelex 100 resin is then added to each tube, and samples are incubated at 56° C for 30 min. This heating step, combined with the alkaline pH of Chelex 100, results in blood cell lysis and denaturation of double-stranded DNA, promoting access and annealing of PCR primers (Walsh et al., 1991). In addition, Chelex 100 resin is postulated to bind metal ion cofactors required for activity of DNA metabolizing enzymes, thus increasing recovery of intact template for PCR, a critical feature in the context of small sample size. The resin/blood cell mixture is vigorously vortexed for 10 sec, and then suspended in boiling water for 8 min. Boiled samples are vortexed for 10 sec., centrifuged for 3 min at 15,000 × g to collect the resin beads at the bottom of the tube. 2.5 mcl of each resulting supernatant is used as a template for PCR as described below. A short protocol summary of this procedure is presented as Table 1.
Table 1.
Genomic DNA Preparation Short Protocol
| Step | Description | Notes |
|---|---|---|
| 1 | Capture bird, lance vessel above knee with sterile needle | See Fig. 2 |
| 2 | Blot blood with sterile chad of 3MM paper | Chad ≈3 mm2 |
| 3 | Place blood sample in sterile 1.5 mL centrifuge tube | |
| 4 | Add 1 mL of sterile water, mix by inversion | |
| 5 | Incubate at RT for 15 min | To allow elution of blood cells from paper |
| 6 | Centrifuge at 15,000 × g for 3 min | To collect blood cells |
| 7 | Carefully remove supernatant leaving ≈20 mcL | Avoid disturbing invisible cell pellet, leave paper in tube |
| 8 | Add 200 mcL of 5% Chelex resin suspension to pellet | |
| 9 | Incubate at 56° C for 15 min | |
| 10 | Mix vigorously by vortex for 5 sec | |
| 11 | Boil for 8 min | |
| 12 | Mix vigorously by vortex for 5 sec | |
| 13 | Collect resin by centrifuging at 15,000 × g for 3 min | |
| 14 | Use supernatant for PCR | Avoid disturbing resin pellet |
| 15 | Store at −20 C | To reuse repeat steps 12 – 14 |
PCR Primer Design
Polymerase Chain Reaction
PCR is done in a total volume of 25 mcl consisting of 50 mM Tris-HCl (pH 7.4), 2.5 mM MgCl2, 200 mcM of each nucleotide, 1.0 mcM of each primer, 0.75 U Taq polymerase and 2.5 mcl of genomic DNA prepared as described above.
To amplify the female-specific W chromosome, the following forward primer was designed and designated W1: GGGTTTTGACTGACTAACTGATT. This was paired with the reverse primer designated W2: GTTCAAAGCTACATGAATAAACA. The size of the fragment of the CHD gene amplified by the combination of W1 and W2 primers is 179 bp. To amplify a fragment of the CHD gene from the Z chromosome of both male and female genomic DNA the following forward primer was designed and designated Z1: GTGTAGTCCGCTGCTTTTGG. This was paired with a reverse primer designated Z2: GTTCGTGGTCTTCCACGTTT. The size of the fragment of the CHD gene amplified from both male and female Z DNA by the combination of Z1 and Z2 primers is 242 bp. Relative migration through a 1.2% agarose gel of products amplified from male and female genomic DNA by each primer set is presented as Fig 1.
We routinely prepare and use a mastermix that includes all PCR components except for the genomic DNA template. This mastermix is stored frozen (−20 C) in aliquots of convenient volume (e.g. 200 mcl). A recipe is presented in Table 2.
Table 2.
Zebra Finch Sex by PCR Mastermix Preparation
| Component | Stock Concentration | Volume to Add | Final Concentration |
|---|---|---|---|
| PCR Buffer1 | 10 X | 100 mcL | 1 X |
| MgCl2 | 25 mM | 100 mcL | 2.5 mM |
| dNTP mix | 10 mM each | 20 mcL | 200 mcM each |
| W1 Primer | 100 mcM | 100 mcL | 1 mcM |
| W2 Primer | 100 mcM | 100 mcL | 1 mcM |
| Z1 Primer | 100 mcM | 100 mcL | 1 mcM |
| Z2 Primer | 100 mcM | 100 mcL | 1 mcM |
| Sterile H2O | --- | 374 mcL | QS to 1000 mcL |
| Taq DNA Polymerase2 | 5 U/mcL | 6 mcL | 0.75 U/22.5 mcL |
500 mM KCl, 100 mM Tris-Cl (pH = 8.3)
Vortex mix other components before adding Taq. Mix Taq by inversion.
After preparation dispense into aliquots and store at −20 C.
The following reaction conditions are employed for a total of 30 cycles using a Bio-Rad iCycler thermalcycler: 94° C for 30 sec, 56° C for 45 sec and 72° C for 45 sec.
Results
Isolation of Genomic DNA
The adapted forensic method has resulted in consistent isolation of genomic DNA suitable for PCR amplification. The most important features of this approach are: (1) that sample collection is quickly completed, minimizing stress to the animal: (2) it requires a very small blood sample yet is reliable and finally; (3) it has not resulted in increased incidence of infection or other potential problems.
Sex Determination by PCR
Use of primer sets specifically targeting zebra finch sex chromosomes has resulted in reliable and unbiased amplification from both Z and W templates (see Fig 1). The method is convenient enough that all birds hatched in our breeding aviary can be sexed prior to experimental assignment.
Discussion
The combination of forensics methods of genomic DNA isolation from very small blood samples with PCR amplification of sex-specific sequences is novel in that it provides a minimally-invasive approach to the problem of sex determination of fledglings. This is a critical feature in the context of sexing very young birds that are fragile and stress-sensitive and represents an important addition to previously reported sexing methods (Griffiths et al., 1998; Runciman et al., 1999).
Approaches similar to the one presented here for zebra finches should be useful for the early sexing of animals in which XY or ZW sex-determination systems are found (Grutzner et al., 2004). This should improve the ability to use these animals in developmental experiments before sex characteristics become apparent.
Prior to development of the sexing methods described here, we could not reliably determine the sex of young zebra finches prior to their assignment to treatment groups. Therefore, both male and female fledglings were initially assigned, effectively doubling the number of subjects required early on. Females were removed from studies following maturation. The need to initially include females in experiments significantly increased effort and consumption of supplies, unbalanced numbers of males across treatment groups, and increased experimental impact on vertebrate animals in a manner that was not productive in-terms of data generation. The reliable means of zebra finch sex determination presented here allows us to avoid these problem and to more efficiently use this species that, due to late-postnatal vocal learning, is particularly useful for neurodevelopmental experiments.
Acknowledgments
This work was supported by NIDA grant 1 R21 DA14693.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- de Kloet RS, de Kloet SR. Evolution of the spindlin gene in birds: independent cessation of the recombination of sex chromosomes at the spindlin locus in neognathous birds and tinamous, a palaeognathous avian family. Genetica. 2003;119:333–42. doi: 10.1023/b:gene.0000003842.72339.df. [DOI] [PubMed] [Google Scholar]
- Griffiths R, Double MC, Orr K, Dawson RJ. A DNA test to sex most birds. Mol Ecol. 1998;7:1071–5. doi: 10.1046/j.1365-294x.1998.00389.x. [DOI] [PubMed] [Google Scholar]
- Grutzner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O'Brien PC, Jones RC, Ferguson-Smith MA, Marshall Graves JA. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004;432:913–7. doi: 10.1038/nature03021. [DOI] [PubMed] [Google Scholar]
- Runciman D, Zann RA, Murray ND. A W-chromosome linked marker for gender identification in the zebra finch. Anim Genet. 1999;30:66–7. doi: 10.1046/j.1365-2052.1999.00323-1.x. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Johnson F. Cannabinoid exposure alters learning of zebra finch vocal patterns. Brain Res Dev Brain Res. 2003;142:215–7. doi: 10.1016/s0165-3806(03)00061-0. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Johnson F. CB1 cannabinoid receptor expression in brain regions associated with zebra finch song control. Brain Research. 2000;857:151–7. doi: 10.1016/s0006-8993(99)02393-8. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Johnson F. The Zebra Finch CB1 Cannabinoid Receptor: Pharmacology and In Vivo and In Vitro Effects of Activation. J Pharmacol Exp Ther. 2001;297:189–97. [PubMed] [Google Scholar]
- Soderstrom K, Tian Q. Developmental Pattern of CB1 Cannabinoid Receptor Immunoreactivity in Brain Regions Important to Zebra Finch (Taeniopygia guttata) Song Learning and Control. J Comp Neurol. 2006;496:739–58. doi: 10.1002/cne.20963. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Tian Q. Distinct Periods of Cannabinoid Sensitivity During Zebra Finch Vocal Development. Developmental Brain Research. 2004;153:225–32. doi: 10.1016/j.devbrainres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Tian Q, Valenti M, Di Marzo V. Endocannabinoids link feeding state and auditory perception-related gene expression. J Neurosci. 2004;24:10013–21. doi: 10.1523/JNEUROSCI.3298-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–13. [PubMed] [Google Scholar]


