Abstract
The functions of neurotransmitters in fetal development are poorly understood. Genetic observations have suggested a role for the inhibitory amino acid neurotransmitter γ-aminobutyric acid (GABA) in the normal development of the mouse palate. Mice homozygous for mutations in the β-3 GABAA receptor subunit develop a cleft secondary palate. GABA, the ligand for this receptor, is synthesized by the enzyme glutamic acid decarboxylase. We have disrupted one of the two mouse Gad genes by gene targeting and also find defects in the formation of the palate. The striking similarity in phenotype between the receptor and ligand mutations clearly demonstrates a role for GABA signaling in normal palate development.
The neurotransmitter γ-aminobutyric acid (GABA) has many critical functions as an intercellular signaling molecule in the nervous system and is thought to be involved in cell signaling in a number of nonneuronal cell types (1). In addition, a role for GABA has been suggested in the development and differentiation of the nervous system (2–4). In the central nervous system, GABA is the primary inhibitory neurotransmitter and has a critical role in the regulation of neural activity. Outside of the nervous system a number of cell types synthesize GABA. For example, GABA is made by the pancreatic β cells and appears to act as a signaling molecule between cells within the pancreatic islets (5, 6). This β cell-specific expression of GABA and glutamic acid decarboxylase (GAD) underlies the destruction of these cell types in autoimmune insulin-dependent diabetes by GAD autoantibodies (7). Several observations have also suggested that GABA plays a role in normal embryonic and fetal development. Cell culture studies have demonstrated that GABA can promote the survival, differentiation, and migration of embryonic neurons (2, 4, 8). In addition, both genetic and teratological studies have shown that GABA signaling may be involved in normal craniofacial development (9–14).
To further study the roles of GABA in normal development and physiology, we have taken a genetic approach. This approach is necessary due to the current lack of specific inhibitors of GAD enzyme activity (15). In mice, two distinct GAD enzymes are encoded by two separate genes, Gad65 and Gad67 (16). We have used gene targeting to inactivate Gad67 and find that the homozygous mutants exhibit a developmental phenotype characterized by neonatal death and a highly penetrant cleft secondary palate. Previous observations have suggested a role for GABA in palate development. Some of these earlier studies showed that drugs potentiating GABA action can induce cleft palate during a critical period of mouse palate development (12–14). However, these studies used high drug doses to produce the cleft palate phenotype, suggesting that the effect might be nonspecific. The analysis of mice with mutations in the β-3 GABAA receptor demonstrated that these mutations are associated with cleft secondary palate in mice (9–11). The phenotype in these receptor mutants showed that this gene is somehow involved in palate development but did not demonstrate that GABA was the ligand involved in this particular function. Our results strengthen and extend the previous studies by clearly demonstrating that GABA has a critical role in the normal development of the mouse palate.
MATERIALS AND METHODS
Construction of the Gad67 Mutant Mice and Histological Analysis.
A Gad67 genomic clone was isolated from an embryonic stem (ES) cell genomic library. The targeting vector was constructed by inserting the pMC1Neo poly(A) gene (17) into a BamHI site in the first protein coding exon of the Gad67 gene (18). The herpes simplex virus (HSV) 1 and HSV2 thymidine kinase genes were added to this construct to provide negative selection. R1 ES cells (19) were electroporated with the Gad67 targeting vector and clones doubly resistant to G418 and 1-(2-deoxy-2-fluoro-β-d-arabinfuranosyl)-5-iodouracil (FIAU) were selected. Of 129 double-resistant ES cell colonies, 9 contained a targeted disruption of the Gad67 gene as determined by Southern blot analysis. Two of these clones were injected into C57BL/6 host blastocyst embryos to generate chimeras. Several of these chimeras passed the Gad67 mutation to their offspring, as determined by Southern blot or PCR analysis. These mice were intercrossed to produce Gad67 homozygotes and control littermates. The Gad67 mutation is maintained on a hybrid 129/B6 genetic background.
Whole-mount skeleton preparations were made as described (20). Fetal and newborn mice were fixed in 4% formaldehyde/PBS and dissected to examine the palates. Gestation time was determined with the assumption that noon of the day a plug was found equals 0.5 day of development.
RNase Protection Analysis.
A probe corresponding to nucleotides 33–253 of the cDNA sequence of Katarova et al. (18) was hybridized to 5 μg of adult brain total RNA or 25 μg of total RNA from embryonic day (E) 18.5 day fetal heads. This probe corresponds to the region of the Gad67 mRNA that is disrupted by the Neo cassette in the mutant allele. The Gad65 probe was as described (21). The GAPDH probe was purchased from Ambion (Austin, TX).
RESULTS AND DISCUSSION
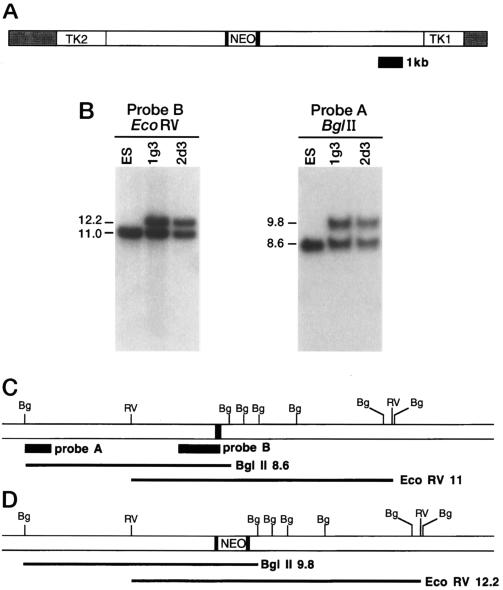
Mice with a loss-of-function mutation in the Gad67 gene were generated by gene targeting in embryonic stem cells (Fig. 1). The homologous recombination event inserted a neo cassette into the first protein-coding exon of the Gad67 gene in ES cells (Fig. 1 B–D). These ES cells were injected into host embryos to generate chimeric mice and several chimeras passed the mutation to their progeny, generating a colony of mice heterozygous for the Gad67 mutation.
Figure 1.
Gene targeting at the Gad67 locus. (A) The Gad67 targeting vector. The pMC1NeoPoly(A) cassette (NEO) was inserted into the first coding exon of the Gad67 gene (solid boxes). HSV1 and HSV2 thymidine kinase genes (TK1 and TK2) were added flanking the homology. The shaded region is plasmid vector sequence. (B) Southern blot analysis of gene targeting at Gad67. Genomic DNA was isolated from the parental ES cell line and from the ES cell lines 1g3 and 2d3 containing the disrupted Gad67 allele. DNA was digested with the indicated restriction enzymes (BglII and EcoRV) and probed with a flanking probe (probe A) and an internal probe (probe B). (C and D) Restriction maps of the wild-type (C) and mutant (D) Gad67 genes. The probes used in the Southern blot analysis and the sizes of the bands detected are indicated below the maps.
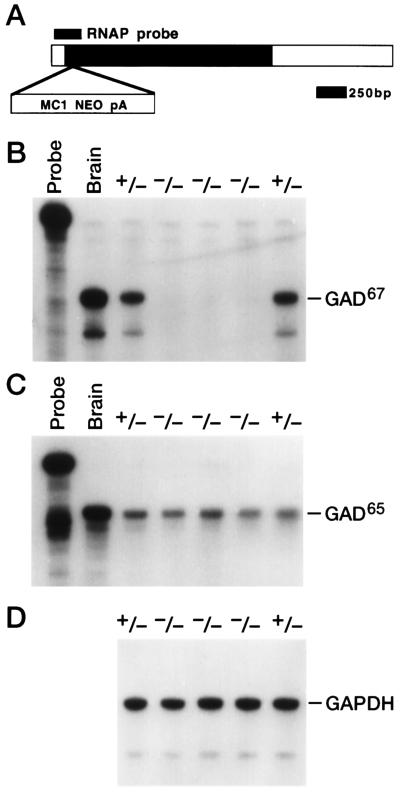
The Gad67 heterozygotes were intercrossed and Gad gene expression was examined in the progeny. In mice, two distinct GADs are encoded by two separate genes, Gad67 and Gad65 (16). We determined the effect of the Gad67 mutation on expression of Gad67 and Gad65 mRNA in mouse fetuses. In an RNase protection assay, a probe spanning the site of the neo insertion did not protect any intact Gad67 transcripts in RNA from Gad67 (−/−) E18.5 mouse heads (Fig. 2B). An RNase protection analysis of the other Gad gene, Gad65, was performed to detect any compensatory changes in its expression in the mutants. Gad65 expression was not altered in the Gad67 homozygotes, showing that no compensatory mechanism exists to alter its RNA expression level in the Gad67 mutants (Fig. 2C). This analysis indicates that the disruption of the Gad67 locus eliminates the expression of intact Gad67 RNA without altering the level of Gad65 mRNA expression.
Figure 2.
RNase protection analysis of the Gad67 and Gad65 genes in Gad67 mutants. (A) The position of the Gad67 RNase protection probe (RNAP probe) is shown relative to the published cDNA sequence. The shaded portion of the cDNA corresponds to the Gad67 coding sequence. The position of the Neo insertion into the coding sequence in the Gad67 mutant allele is indicated (MC1NEOpA). (B–D) RNase protection of Gad67 RNA. Total RNA (25 μg) from individual E18.5 day mouse heads of the indicated genotype (heterozygote +/+ and homozygous mutant −/−) and 5 μg of adult brain RNA (brain) were subjected to RNase protection analysis with the Gad67 probe (B), the Gad65 probe (C), and a control GAPDH probe (D). Undigested probe (Probe) is shown for the Gad65 and Gad67 assays (B and C).
In addition to Gad gene expression, the viability and outward phenotype of the Gad67 mutants was examined. The Gad67 homozygous mutants die shortly after birth. Genotype analysis of the progeny of Gad67 heterozygote intercrosses shows that no homozygotes were recovered at weaning (Table 1). However, nearly expected numbers of homozygotes were recovered shortly after birth and normal numbers were found in litters removed by caesarean section late in gestation (Table 1). Gad67 (−/−) E18.5 day fetuses, delivered by caesarean section, were always distinguishable from their littermates, retaining after delivery a blue skin color suggesting hypoxia. Newborn homozygotes that survived birth failed to nurse, leading to fatal dehydration and frequently aspirated air into their intestinal tract. Bloating of the intestinal tract is observed in other mutant mouse strains with cleft palates (22). A preliminary histological examination of two Gad67 homozygotes revealed no gross defects in brain morphology. A more detailed examination of nervous system anatomy and function in these mice is underway.
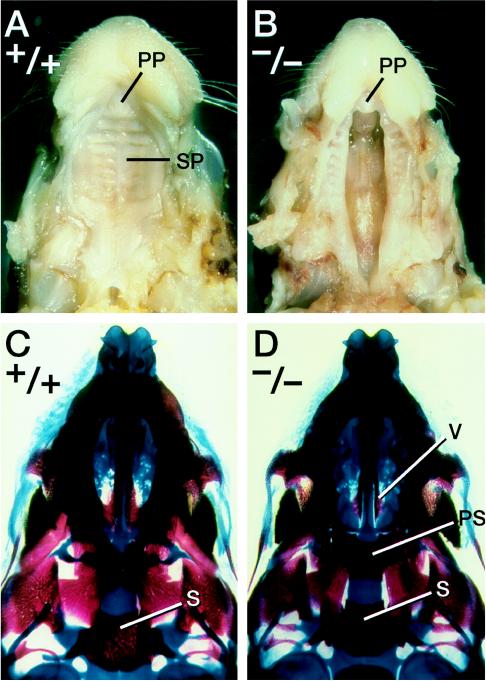
The cleft palate phenotype found in the β-3 GABAA receptor subunit mutants (10, 11) prompted a careful examination of the palates in Gad67 mutants. The palates of 25 Gad67 homozygous mutants were examined. Overall, the cleft palate was observed in 92% (23 of 25) of the homozygotes and was absent in all of the heterozygous and wild-type progeny. The cleft palate phenotype observed in the Gad67 homozygotes is always a complete cleft of the secondary palate (Fig. 3 A and B). In these homozygotes, the cleft of the palatal shelves is wide, clearly revealing the bones of the nasal cavity (Fig. 3 B and D). Coronal sections of Gad67 (−/−) newborns and E18.5 day fetuses show that the palatal shelves did elevate but had failed to fuse (data not shown). In the Gad67 mutants, some narrowing of the maxilla was observed; however, no defects in the other bones and cartilages derived from the first pharyngeal arch were found (Fig. 3 and data not shown). In particular, the mandible is of normal size and morphology. No defects were found in the hyoid bone or in the thyroid and cricoid cartilages that are derived from the more posterior pharyngeal arches. This analysis shows that the craniofacial defect in the Gad67 (−/−) mutants is a specific defect in palatogenesis.
Figure 3.
Cleft palate phenotype in Gad67 mutants. (A and B) Palatal views of a wild-type newborn (A) and its homozygous mutant littermate (B). The primary palate (PP) and secondary palate (SP) are indicated. (C and D) Palatal view of cleared skeletal preparations of a wild-type newborn (C) and a Gad67 homozygous mutant (D). In the mutant, the vomer (V) and presphenoid (PS) are visible due to the cleft secondary palate. s, Sphenoid bone.
The cleft palate phenotype in mice mutant for the GABA-synthesizing enzyme Gad67 indicates a role for GABA function in the development of the palate. The striking similarity between the cleft palate seen in the Gad67 (−/−) mice and the defect reported in the β-3 GABAA receptor subunit mutant suggests that GABA signaling through GABAA receptors is required for normal development of the mouse palate. Consistent with a direct role of GABA signaling in palatogenesis, GABA has been detected in the developing mouse palate (12).
In addition to genetic observations, it has been shown that drugs altering GABA signaling can induce cleft palate in developing mice. The benzodiazepine drug diazepam can induce cleft palate when administered to pregnant mice during a critical period of palatogenesis (13, 14). Diazepam potentiates GABA activation of GABAA receptors, increasing GABA function. The induction of cleft palate both by loss-of-function mutations and by an enhancement of function by benzodiazepines suggests that a process during palatogenesis requires a specific range of GABA signaling for normal development. In humans, a link between benzodiazepine use during pregnancy and craniofacial defects has been reported (23–25) but is controversial (26). The highly penetrant cleft palate phenotype seen in our mice and in the GABAA receptor knockout indicates that the role of benzodiazepines in human craniofacial defects should be carefully reexamined given the widespread use of these drugs.
In humans, isolated cleft palate is common and has a complex multifactorial etiology (27). At present, no human syndromes or disorders that feature cleft palate map near the Gad67 gene. In mice, Gad67 is closely linked to two other mutations that cause craniofacial defects including cleft palate, the spontaneous mutation First arch (Far) and a targeted disruption of the homeobox gene Dlx-2. These genes map near Gad67, very close to the HoxD complex on chromosome 2. In contrast to the phenotypes of the Dlx-2 and Far mutants, the effect of the Gad67 mutation is specific to development of the palate with no alterations in other craniofacial structures (22, 28). It is notable that this 6-centimorgan region of chromosome 2 contains a high density of genes involved in development, including craniofacial formation and palatogenesis. The cleft palate phenotype in mice mutant for Gad67 or for the β-3 subunit of the GABAA receptor suggests that genetic or environmental perturbations of GABA pathways during fetal development can alter palatogenesis. Genes encoding the enzymes of GABA metabolism and those encoding GABA receptors should be considered as candidates for loci involved in the etiology of human cleft palate.
Gene targeting has also been used to inactivate the Gad65 gene in mice (S. Kash and S. Baekkeskov, personal communication). Unlike Gad67 mutants, Gad65 homozygotes are viable but exhibit temporal lobe epilepsy. The availability of mice deficient in both Gad loci will allow tests of synergistic interactions and functional redundancies between the two genes. It will be of interest to examine craniofacial development and the neuroanatomy of Gad double mutants. In addition, new seizure phenotypes may be revealed in Gad65 (+/−)/Gad67 (+/−) and Gad65 (−/−)/Gad67 (+/−) mice, providing further insight into the genetic mechanisms of epilepsy.
Table 1.
Gad67 intercross progency genotypes
| Stage | +/+ | +/− | −/− |
|---|---|---|---|
| Late gestation | 21 | 39 | 22 |
| Newborn | 24 | 42 | 12 |
| Adult | 28 | 43 | 0 |
Late gestation fetuses were removed by caesarean section on E16.5–E18.5. Adults were genotyped at 4–6 weeks old. Numbers of animals with a specific genotype are shown.
Acknowledgments
We thank C. Lenz, M. Allen, G. Peterson, E. Nakashima, M. Wagstaff, and S. Barnett for excellent technical assistance and L. Oswald for preparation of the manuscript.
ABBREVIATIONS
- GABA
γ-aminobutyric acid
- GAD
glutamic acid decarboxylase
- ES cell
embryonic stem cell
- HSV
herpes simplex virus
- E
embryonic day
References
- 1.Erdo S L, Wolff J R. J Neurochem. 1990;54:363–372. doi: 10.1111/j.1471-4159.1990.tb01882.x. [DOI] [PubMed] [Google Scholar]
- 2.Barbin G, Pollard H, Gaiarsa J L, Ben-Ari Y. Neurosci Lett. 1993;152:150–154. doi: 10.1016/0304-3940(93)90505-f. [DOI] [PubMed] [Google Scholar]
- 3.LoTurco J L, Owens D F, Heath M J S, Davis M B E, Kriegstein A R. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 4.Behar T N, Yong-Xin L, Tran H T, Ma W, Dunlap V, Scott C, Barker J L. J Neurosci. 1996;16:1808–1818. doi: 10.1523/JNEUROSCI.16-05-01808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rorsman P, Berggren P O, Bokvist K, Ericson H, Mohler H, Ostnson C G, Smith P A. Nature (London) 1989;341:233–236. doi: 10.1038/341233a0. [DOI] [PubMed] [Google Scholar]
- 6.Reetz A, Solimena M, Matteoli M, Folli F, Takei K, De Camilli P. EMBO J. 1991;10:1275–1284. doi: 10.1002/j.1460-2075.1991.tb08069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baekkeskov S, Aanstoot H-J, Christgau S, Reetz A, Solimena M, Cascalho M, Folli F, Richter-Oleson H, DeCamilli P. Nature (London) 1990;347:151–156. doi: 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Morrow A L, Devaud L, Grayson D R, Lauder J M. J Neurosci. 1997;17:2420–2428. doi: 10.1523/JNEUROSCI.17-07-02420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culiat C T, Stubbs L, Nicholls R D, Montgomery C S, Russell L B, Johnson D K, Rinchik E M. Proc Natl Acad Sci USA. 1993;90:5105–5109. doi: 10.1073/pnas.90.11.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Culiat C T, Stubbs L J, Woychik R P, Russell L B, Johnson D K, Rinchik E M. Nat Genet. 1995;11:344–346. doi: 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- 11.Homanics G E, DeLorey T M, Firestone L L, Quinlan J J, Handforth A, Harrison N L, Krasowski M D, Rick C E M, Korpi E R, Makela R, Brilliant M H, Hagiwara N, Ferguson C, Snyder K, Olsen R W. Proc Natl Acad Sci USA. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmerman E F, Wee E L. Curr Top Dev Biol. 1984;19:37–63. doi: 10.1016/s0070-2153(08)60394-4. [DOI] [PubMed] [Google Scholar]
- 13.Miller R P, Becker B A. Toxicol Appl Pharmacol. 1975;32:53–61. doi: 10.1016/0041-008x(75)90194-5. [DOI] [PubMed] [Google Scholar]
- 14.Wee E L, Zimmerman E F. Teratology. 1983;28:15–22. doi: 10.1002/tera.1420280104. [DOI] [PubMed] [Google Scholar]
- 15.Cooper J R, Bloom F E, Roth R H. The Biochemical Basis of Neuropharmacology. Oxford, U.K.: Oxford Univ. Press; 1996. [Google Scholar]
- 16.Erlander M G, Tillakaratne N J K, Feldblum S, Patel N, Tobin A J. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- 17.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 18.Katarova Z, Szabo G, Mugnaini E, Greenspan R J. Eur J Neurosci. 1990;2:190–202. doi: 10.1111/j.1460-9568.1990.tb00412.x. [DOI] [PubMed] [Google Scholar]
- 19.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Condie B G, Capecchi M R. Development (Cambridge, UK) 1993;119:579–595. doi: 10.1242/dev.119.3.579. [DOI] [PubMed] [Google Scholar]
- 21.Bain G, Ramkumar T P, Cheng J M, Gottlieb D I. Mol Brain Res. 1993;17:23–30. doi: 10.1016/0169-328x(93)90068-z. [DOI] [PubMed] [Google Scholar]
- 22.Qiu M, Bulfone A, Martinez S, Meneses J J, Shimamura K, Pederson R A, Rubenstein J L R. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 23.Saxen I, Saxen L. Lancet. 1975;ii:498. doi: 10.1016/s0140-6736(75)90567-x. [DOI] [PubMed] [Google Scholar]
- 24.Aarskog D. Lancet. 1975;ii:921. doi: 10.1016/s0140-6736(75)92153-4. [DOI] [PubMed] [Google Scholar]
- 25.Safra M J, Oakley G P. Lancet. 1975;ii:478–480. doi: 10.1016/s0140-6736(75)90548-6. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg L, Mitchell A A, Parsells J L, Pashayan H, Louik C, Shapiro S. N Engl J Med. 1983;309:1282–1285. doi: 10.1056/NEJM198311243092103. [DOI] [PubMed] [Google Scholar]
- 27.Moore G, Ivens A, Chambers J, Bjornsson A, Arnason A, Jensson O, Williamson R. Development (Cambridge, UK) Suppl. 1988;103:233–239. doi: 10.1242/dev.103.Supplement.233. [DOI] [PubMed] [Google Scholar]
- 28.Juriloff D M, Harris M J. J Hered. 1991;82:402–405. doi: 10.1093/oxfordjournals.jhered.a111111. [DOI] [PubMed] [Google Scholar]