SUMMARY
Set/template-activating factor (TAF)-Iβ, part of the Set-Can oncogene product found in acute undifferentiated leukemia, is a component of the inhibitor of acetyltransferases (INHAT) complex. Set/TAF-Iβ interacted with the DNA-binding domain of the glucocorticoid receptor (GR) in yeast two-hybrid screening, and repressed GR-induced transcriptional activity of a chromatin-integrated glucocorticoid-responsive and a natural promoter. Set/TAF-Iβ was co-precipitated with glucocorticoid response elements (GREs) of these promoters in the absence of dexamethasone, while addition of the hormone caused dissociation of Set/TAF-Iβ from and attraction of the p160-type coactivator GRIP1 to the promoter GREs. Set-Can fusion protein, on the other hand, did not interact with GR, was constitutively co-precipitated with GREs and suppressed GRIP1-induced enhancement of GR transcriptional activity and histone acetylation. Thus, Set/TAF-Iβ acts as a ligand-activated GR-responsive transcriptional repressor, while Set-Can does not retain physiologic responsiveness to ligand-bound GR, possibly contributing to the poor responsiveness of Set-Can-harboring leukemic cells to glucocorticoids.
Keywords: glucocorticoid receptor, Set/TAF-Iβ, inhibitor of histone acetyltransferases complex, histone acetylation, set-can translocation, and leukemia
INTRODUCTION
Glucocorticoids play an essential physiologic role in the regulation of basal and stress-related homeostasis [1]. At “pharmacologic” doses, glucocorticoids are an indispensable therapy for many inflammatory, autoimmune, allergic and lymphoproliferative diseases, acting as potent immunosuppressive, anti-inflammatory and pro-apoptotic agents [2]. This major physiologic/pharmacologic importance of glucocorticoids suggests that insensitivity of tissues to glucocorticoids may influence their physiologic actions, as well as the course of pathologic states [3]. Indeed, several autoimmune/allergic/inflammatory diseases, such as rheumatoid arthritis, bronchial asthma and Crohn’s disease, and lymphoproliferative diseases, including acute lymphocytic leukemia and malignant lymphoma, develop glucocorticoid resistance in immune or malignant cells/tissues, respectively, which reduces the efficacy of glucocorticoid therapy [3,4].
The actions of glucocorticoids are mediated by the ubiquitous intracellular glucocorticoid receptor (GR), which functions as a hormone-activated transcription factor of glucocorticoid-target genes [5]. The GR consists of three domains, the N-terminal or “immunogenic” domain, the central, DNA-binding domain (DBD), and the C-terminal, ligand-binding domain (LBD). Ligand-activated GR translocates into the nucleus, binds to the glucocorticoid response elements (GREs) and attracts several so-called coactivators and chromatin-remodeling factors to the promoter region of glucocorticoid-responsive genes through its two transactivation domains activation function (AF) 1 and AF2 [5]. One of such protein groups, the histone acetyltransferase (HAT) coactivators, acetylate specific lysine residues located in the N-terminal tail of chromatin-bound histones, and facilitate access of other transcription factors and transcriptional machineries to the promoter region [6]. Among them, the p160 type HAT coactivators, like the steroid receptor coactivator 1 (SRC1) and the glucocorticoid receptor-interacting protein 1 (GRIP1), play an essential role in GR-induced transcriptional activity, being attracted to the promoter region at an early phase of the transcriptional process [6].
In contrast to coactivators, corepressors of transcription, such as the nuclear receptor corepressor (NCoR) and the silencing mediator for retinoid and thyroid hormone receptors (SMRT), attract histone deacetylases (HDACs) to the promoter region and silence transcription by deacetylating histones [6,7]. We recently found that Smad6, a regulatory molecule downstream of the transforming growth factor β receptor signaling, attracted HDAC3 to GR-bound promoters and repressed glucocorticoid-stimulated transcription by preventing and/or reversing the acetylation caused by the p160 coactivators [8].
The inhibitor of histone acetyltransferases (INHAT) complex, a trimer consisting of template activating factor I (TAF-I) α, Set/TAF-Iβ and pp32, binds lysine residues of histones and protects them from acetylation promoted by HAT-bearing transcription factor or nuclear receptor coactivators [9]. The human TAF-Iα and Set/TAF-Iβ, both ubiquitously expressed proteins and members of a large family of histone chaperones, share the same 277 amino acid sequence except a 13 amino acid insertion in the N-terminal portion of TAF-Iα [10]. Set/TAF-Iβ was originally found in acute undifferentiated leukemia cells as part of a fusion oncoprotein containing Can that is a nucleoporin involved in nucleocytoplasmic transport of protein and mRNA [11–13]. Set/TAF-Iβ has multiple distinct activities, such as inhibition of phosphatase 2A activity, induction of cellular transformation and differentiation, and transfer of histones onto naked DNA [14–20]. The INHAT complex suppresses the transcriptional activity of several transcription factors and nuclear receptors, such as Sp1, the Kruppel-like factor (KLF) 5 and the estrogen receptor (ER) α [21–23]. The INHAT complex interacts with the DBD of these trans-acting factors through Set/TAF-Iβ, but the physiologic importance of the interaction between the INHAT complex and these transcription factors remains to be determined [21–23]. It is known, however, that Set/TAF-Iβ facilitates the association of ERα with its DNA response elements and is required for the transcription of chromatin templates [17,23]. These data indicate that Set/TAF-Iβ might also function as a positive regulator of ERα-driven transcription, in addition to exerting its principally inhibitory effect.
To identify potential factors that contribute to the development of glucocorticoid insensitivity observed in pathologic conditions, we performed yeast two-hybrid screening assays using the GR DBD as bait. We found Set/TAF-Iβ, a component of the INHAT complex, as a specific GR DBD interactor. Set/TAF-Iβ already associated with histones interacted with GR in a ligand-dependent fashion and suppressed GR-induced transcriptional activity. Interestingly, ligand-activated GR released the INHAT complex from histone-bound GREs a phenomenon dependent on the physical interaction between the GR DBD and a specific GR-binding domain of Set/TAF-Iβ. Pathologic fusion of Set with Can stayed continuously with the GREs, inhibited acetylation of histone H3 and prevented GRIP1-induced enhancement of GR transcriptional activity. These results indicate that the Set-Can oncoprotein may underlie the glucocorticoid insensitivity observed in patients with acute undifferentiated leukemia with Set-Can translocation [24].
MATERIALS AND METHODS
Plasmids
pRShGRα, pCDNAI/Amp-MR and pSVPRA, which respectively express the human GRα, mineralocorticoid receptor (MR) and progesterone receptor (PR)-A, were generous gifts from Drs. R.M. Evans (Salk Institute, La Jolla, CA), N. Warriar (Centre Recherche Hôtel-Dieu Qué-bec and Laval University, Québec, Canada), and S.S. Simons Jr. (National Institutes of Health, Bethesda, MD). pCHA-TAF-Iα and -TAF-Iβ, which express His-tagged TAF-Iα and Set/TAF-Iβ respectively, were kind gifts from Dr. K. Nagata (Univ. of Tsukuba, Japan) [10]. pSCTOP-Set-Can, which expresses the His-tagged Set-Can fusion protein, was a generous gift from Dr. G. Grosveld (St. Jude Children’s Research Hospital, TN). pcDNA3.1His/B-Set/TAF-Iβ-(2–225), -(2–180), -(181–277), -(181–225), -(225–277), -(Δ181–225) and -Set-Can were constructed by subcloning the coding sequences of the corresponding human Set/TAF-Iβ fragments or Set-Can into pcDNA3.1His/B (Invitrogen, Carlsbad, CA). pRSV-RelA and pRSV-p50, which express p65 and p50 components of the nuclear factor-κB (NF-κB) transcription factor, and (κB)3-Luc, which has 3 κB-responsive elements from the IκB promoter, were described previously [25]. pSG5-GRIP1 was a generous gift from Dr. M.R. Stallcup (University of Southern California, Los Angels, CA). pMMTV-Luc, which expresses the luciferase under the control of the full-length mouse mammary tumor virus (MMTV) promoter that contains four functional GREs [26], was kindly provided by Dr. G.L. Hager (National Institutes of Health, Bethesda, MD). pMAM-neo-Luc, which expresses the luciferase under the control of the full-length MMTV promoter and contains a neomycin-resistant cassette, was purchased from Clontech (Mountain View, CA). pSV40-β-Gal, which expresses β-galactosidase under the control of the Simian virus 40 promoter, was obtained from Promega (Madison, WI).
pLexA-GR-(420–489) was constructed by subcloning the coding sequence of the human GRα DBD (amino acids from 420–489) into pLexA (Clontech). pB42AD-Set/TAF-Iβ-(2–277), -(2–225), -(2–180), -(26–277), -(26–225), -(26–180), -(181–277), -(181–225), -(225–277), -(Δ181–225) and -(2–277) were all constructed by subcloning the coding sequences of the corresponding human Set/TAF-Iβ fragments into pB42AD (Clontech). pOP8-LacZ was purchased from Clontech.
Yeast two-hybrid screening and assay
The yeast two-hybrid screening was performed using GR-(420–489) as bait in the human Jurkat cell cDNA library with the LexA system (Clontech) [27]. For yeast two-hybrid assays, yeast strain EGY48 (Clontech) was transformed with pOP8-LacZ and the indicated pLexA- and pB42AD-based plasmids. The cells were cultured in appropriate selection media and the β-galactosidase activity was measured in the cell suspension as previously described [8]. The β-galactosidase activity was normalized for O.D. value at 600 nm, and fold induction was calculated by the ratio of adjusted β-galactosidase values of transformed cells cultured in the presence of galactose/raffinose vs. those in the medium containing glucose.
Cell cultures and transfection
Human colon carcinoma HCT116 cells were purchased from the American Type Culture Collection (Manassas, VA) and were maintained in McCoy’s 5A medium supplemented with 10% fetal bovine serum, 50 units of penicillin, and 50 μg/ml of streptomycin. HCT116/MMTV cells, which were stably transformed with pMAM-neo-Luc, were maintained in the McCoy’s 5A medium containing 0.2 mg/ml of neomycin and the same supplements [8]. Rat hepatoma HTC cells were maintained in Dulbecco’s Modified Eagle medium with the same amounts of penicillin and streptomycin, and were described previously [28]. HCT116 and HCT116/MMTV cells do not contain functional GR, while HTC cells express the fully active GR.
HCT116 and HCT116/MMTV cells were transfected with Lipofectin® (Invitrogen), as described previously [8]. HCT116 cells were transfected with the indicated amounts of Set/TAF-Iβ- or Set/Can-related plasmids together with 0.5 μg/well of a GR-, MR-, PR-A-, or p65-/p50-expressing plasmid, and 1.0 μg/well of pMMTV-Luc or 0.3 μg/well (κB)3-Luc. 0.5 μg/well of pSV40-β-Gal was always included as an internal control. pMMTV-Luc was omitted in the transfection of HCT116/MMTV cells. Empty vectors were used to maintain the same amounts of transfected DNA. 10−6 M of dexamethasone (Dex) or progesterone (Prog), or 10−8 M of aldosterone (Aldo) (Sigma Chemical Co., St. Louis, MO), was added to the medium 24 hours after transfection. Cell lysates were then harvested after an additional 24 hours, and luciferase and β-galactosidase assays were performed as described previously [29].
Introduction of Set/TAF-Iβ small interfering RNAs (siRNAs) into HTC Cells, the tyrosine aminotransferase (TAT) assay and the real-time PCR
siRNA for the rat Set/TAF-Iβ (5′-GAGACAGAGAGCUUCUUUAdTdT-3′), which targets nucleotides 586–604 of the rat Set/TAF-Iβ coding sequence (GenBank™ accession number XM_233085), was produced by Qiagen (Valencia, CA). The negative control siRNA (5′-UUCUCCGAACGUGUCACGUdTdT-3′) was purchased from Qiagen. HTC cells were transfected with siRNAs by using the Nucleofector system (Amaxa GmbH, Cologne, Germany) with solution-R and protocol-T27, as previously described [28]. We achieved nearly 80% transfection efficiency in these cells with this combination (data not shown). Twenty-four hours after transfecting/plating the cells in 24-well plates, they were stimulated with 10−6 M of dexamethasone. After an additional 24 hours of incubation, cell lysates were obtained for the tyrosine aminotransferase (TAT) assay and the total RNA for the real-time PCR were harvested. TAT assays were performed as previously described [28].
The reverse transcription reaction was carried out as previously described [8]. To detect mRNA levels of the rat TAT, Set/TAF-Iβ and control acidic ribosomal phosphoprotein P0 (RPLP0), primer pairs (TAT, forward: 5′-GTCGCTTCTTACTACCAC-3′, reverse: 5′-CAGGCAGGAGATTGTAGAG-3′; Set/TAF-Iβ, forward: 5′-GAGAGCGGTGACCCATCTTC-3′, reverse: 5′-CCTCTGCCTCTCCTCCTTC-3′; RPLP0, forward: 5′-GACATGCTGCTGGCCAATAAG–3′, reverse: 5′-CAACATGTTCAGCAGTGTG-3′) were used [28]. The real-time PCR was performed in triplicate using the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a 7500 Real-time PCR System (Applied Biosystems), as previously described [8]. Obtained CT (threshold cycle) values of TAT and Set/TAF-Iβ were normalized for those of RPLP0, and their relative mRNA expression was demonstrated as fold induction above the baseline. The dissociation curves of the used primer pairs showed a single peak, and after the PCRs, the samples had a single expected DNA band in an agarose gel analysis (data not shown).
Regular co-immunoprecipitation assays and Western blots of the Set/TAF-Iβ-related molecules and Set-Can
HCT116/MMTV cells were transfected with pcDNA3.1His/B, pcDNA3.1His/B-Set/TAF-Iβ-(2–277), -(Δ181–225) or -Set-Can together with pRShGRα, and were treated with 10−6 M of dexamethasone or vehicle for 2 hours. The cells were then harvested, lysed in buffer containing 50mM Tris-HCl [7.4], 150mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 0.5 M hydroxylamine HCl (PIERCE, Rockford, IL) and 1Tab/50 ml Complete™ Tablet (Roche Applied Science, Indianapolis, IN), and were subsequently centrifuged at 14,000 rpm for 15 min. Co-immunoprecipitation was then carried out with the supernatant as previously described [28]. Proteins were immunoprecipitated with anti-hGRα or control rabbit IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and the protein-antibody complexes were collected with Protein Agarose A/G PLUS (Santa Cruz Biotechnology, Inc.). After cross-linking of proteins was reversed, the samples were separated in SDS-PAGE gels blotted on nitrocellulose membranes. His-Set-TAF-Iβ-related molecules or His-Set-Can were then detected by anti-His antibody (Santa Cruz Biotechnology, Inc.). To evaluate exogenously expressed GR, Set-TAF-Iβ-related molecules or Set-Can, 10% of the cell lysates used in the co-immunoprecipitation reaction were run on SDS-PAGE gels.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed by using the Chromatin Immunoprecipitation Kit (Upstate, Charlottesville, VA) with minor modification in HCT116/MMTV cells, which have integrated in their genome the full-length MMTV promoter driving the luciferase gene, or in HTC cells, which express the endogenous glucocorticoid-responsive TAT [8]. Briefly, 5×106 HCT116/MMTV cells grown in 15 cm-dishes were transfected with 1 μg/well of plasmid expressing the His-tagged full-length, Δ181–225, 181–225 fragment of Set/TAF-Iβ or the His-tagged Set-Can, together with 10 μg/well of pRShGRα. Twenty-four hours after transfection, the cells were exposed to either 10−6 M of dexamethasone or vehicle for 2 hours. 1×106 HTC cells grown in 10 cm-dishes were also exposed to either 10−6 M of dexamethasone or vehicle for 2 hours. They were then fixed with 1% formaldehyde and homogenized, and cross-linked DNA/protein complexes were co-precipitated with anti-His, anti-Set/TAF-Iβ, anti-GRα, anti-GRIP1 (Santa Cruz Biotechnologies, Inc., Santa Cruz, CA) or anti-pp32 (IMEGENEX, San Diego, CA) antibodies, or rabbit control IgG (Santa Cruz Biotechnologies, Inc.). The promoter region -219 to -47 of the MMTV long terminal repeat (fragment size 173 bps), which contains two functional GREs, was amplified from the prepared DNA samples using a primer pair: 5′-AACCTTGCGGTTCCCAG-3′ and 5′-GCATTTACATAAGATTTGG-3′, while tandem endogenous GREs of the rat TAT promoter, which are located ~2,500 bps upstream of its transcription initiation site, were amplified by a primer pair: 5′-TCTTCTCAGTGTTCTCTATCAC-3′ and 5′-CAGAAACCGACAGGCGACTACG-3′ (fragment size 220 bps), as described previously [30,31]. Amplified products were then run on a 2–3% agarose gel, and visualized DNA bands were photographed. For ChIP experiments examining the acetylation of histone H3 bound to GREs, HCT116/MMTV cells were transfected with the indicated plasmids. The human Set/TAF-Iβ siRNA (Santa Cruz Biotechnologies, Inc.) or control siRNA was transfected by using the Nucleofector system (solution-V and protocol-D32) with over 80% transfection efficiency. After cells were treated with dexamethasone and associated DNA was co-precipitated with anti-acetylated histone H3 (K14) antibody (Upstate), the promoter region -219 to -47 of the MMTV long terminal repeat was amplified with the SYBR green real-time PCR by using the primer pair indicated above. Obtained CT (threshold cycle) values of ChIP samples were normalized for those of the corresponding inputs and their relative acetylation of histone H3 (K14) was demonstrated as fold acetylation above the baseline. Total RNA was also purified from the samples in the same experiments, and mRNA levels of the human Set/TAF-Iβ and the control RPLP0 were evaluated with the SYBR green real-time PCR by using the primer pairs (Set/TAF-Iβ: forward: 5′-GTCCACCGAAATCAAATGG-3′, reverse: 5′-CATCAGAATGGTCAGTAAACC-3′; RPLP0: forward: 5′-CGCGACCTGGAAGTCCAACT-3′, reverse: 5′-CCATCAGCACCACAGCCTTC-3′). CT values of the Set/TAF-Iβ mRNA were normalized for those of RPLP0, and relative mRNA expression levels were shown as fold induction above the baseline
Statistical Analysis
Statistical analysis was carried out by ANOVA, followed by Bonferroni correction for multiple comparisons, or unpaired Student t test with the two-tailed p value.
RESULTS
TAF-Iα and Set/TAF-Iβ suppress GR-induced transcriptional activity
Among ~100 clones obtained as interactors of the GR DBD in the LexA-derived yeast two-hybrid screening assay using a Jurkat cell cDNA library, we found two independent clones, which contained the nucleotide sequences corresponding to the C-terminal portion of TAF-I. TAF-I is a component of the INHAT complex, which represses the transcriptional activity of several nuclear hormone receptors [9,32]. Thus, we tested whether over-expressed TAF-Iα or Set/TAF-Iβ, which share the identified interacting fragment, influence GR-induced transcriptional activity from transiently transfected or chromatin-integrated glucocorticoid-responsive MMTV promoter in HCT116 and HCT116/MMTV cells (Figure 1). Both TAF-Iα and Set/TAF-Iβ dose-dependently suppressed the transcriptional activity of the chromatin-incorporated MMTV-promoter in HCT116/MMTV cells (Figure 1A, left panel). Overexpression of Set/TAF-Iβ shifted the dexamethasone titration curve of the luciferase activity downward in these cells (Figure 1A, right panel). These results indicate that TAF-Iα and Set/TAF-Iβ function as negative regulators of GR-induced transcriptional activity.
Figure 1. Set/TAF-Iβ represses GR-induced transcriptional activity.
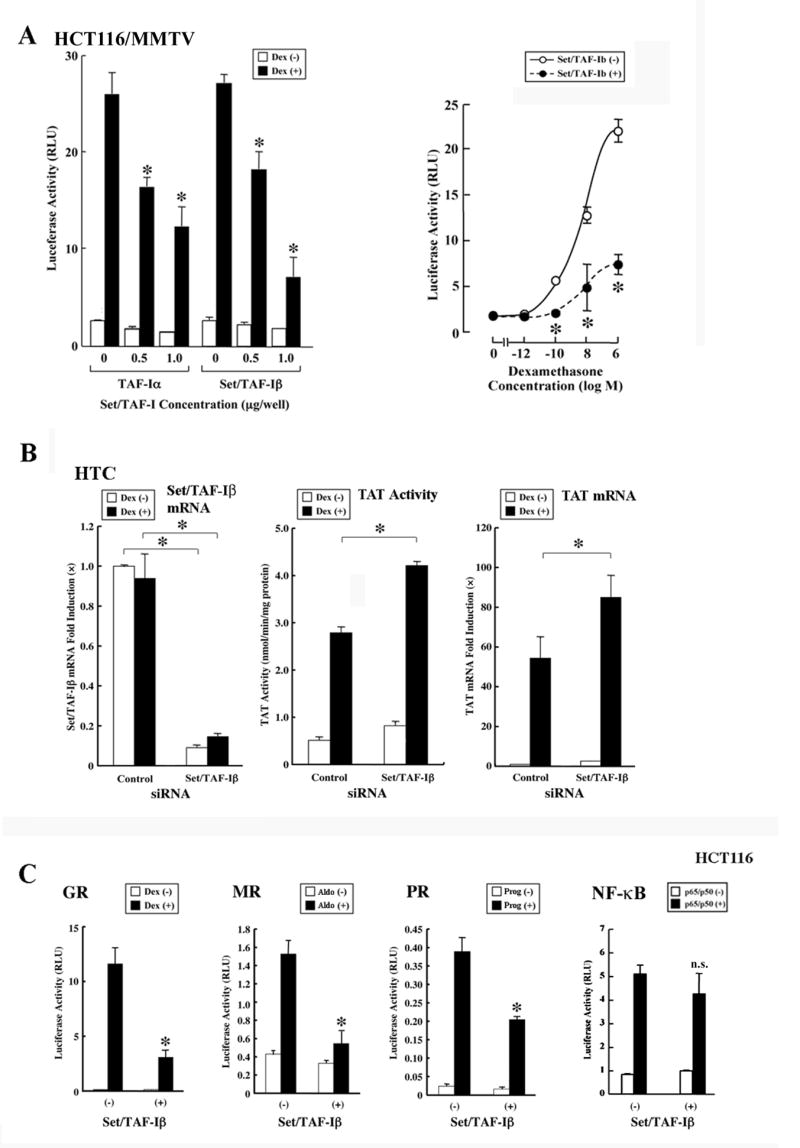
A: TAF-Iα and Set/TAF-Iβ repress GRα-induced transcriptional activity of the MMTV promoter stably incorporated in host cell chromatin in HCT116/MMTV cells.
HCT116/MMTV cells were transfected with the indicated amounts of the TAF-Iα-or Set/TAF-Iβ-expressing plasmid, together with pRShGRα and pSV40-β-Gal.
Left panel: Dose effects of TAF-Iα and Set/TAF-Iβ are shown. Bars represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6 M of dexamethasone (Dex). *: p<0.01 (ANOVA, followed by Bonferroni correction), compared to the baseline (the value obtained in the absence of TAF-Iα and Set/TAF-Iβ, but in the presence of dexamethasone).
Right panel: Set/TAF-Iβ shifts downward the dexamethasone titration curve of luciferase activity from the chromatin-integrated MMTV promoter. Circles represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the indicated concentrations of dexamethasone (Dex). *: p<0.01 (ANOVA, followed by Bonferroni correction), obtained by comparing the results in the absence and presence of Set/TAF-Iβ transfection.
B: Endogenous Set/TAF-Iβ is a negative regulator of GR-induced transcriptional activity in HTC cells.
HTC cells were transfected with Set/TAF-Iβ siRNA or control siRNA. The cells were stimulated with 10−6M of dexamethasone (Dex). Bars indicate the mean ± S.E. of the tyrosine aminotransferase (TAT) activity, mRNA levels of TAT or those of Set/TAF-Iβ. *: p<0.01 (unpaired two-tailed Student t test), by comparing the two values indicated.
C: Set/TAF-Iβ represses the transcriptional activity of the mineralocorticoid (MR) and progesterone receptor (PR)-A in addition to that of the GR.
HCT116 cells were transfected with Set/TAF-Iβ-expressing plasmid together with GRα-, MR- or PR-A-expressing plasmid, pMMTV-Luc or (κB)3-Luc, and pSV40-β-Gal. Bars represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6 M of dexamethasone (Dex) or progesterone (Prog), 10−8 M of aldosterone (Aldo), or p65/p50 transfection. *: p<0.01, n.s.: not significant (unpaired two-tailed Student t test), compared to the baseline (“(-)” in the presence of ligand or p65/p50).
We examined the effects of depletion of endogenous Set/TAF-Iβ on the expression of the glucocorticoid-responsive TAT gene by using siRNA for Set/TAF-Iβ in rat HTC cells (Figure 1B). The TAT gene is well-known glucocorticoid-responsive gene, whose responsiveness is dependent on tandem GREs located ~2,500 bps upstream of its transcription initiation site [33]. Transfection of Set/TAF-Iβ siRNA strongly suppressed the mRNA expression from the endogenous Set/TAF-Iβ gene and enhanced both TAT enzymatic activity and its mRNA expression in these cells. Thus, endogenous Set/TAF-Iβ functions as a negative regulator of the GR transcriptional activity on an endogenous glucocorticoid-responsive gene.
We further examined Set/TAF-Iβ for a potential effect on steroid hormone receptors MR and PR-A in addition to GR in HCT116 cells (Figure 1C). Set/TAF-Iβ suppressed the transcriptional activity of these receptors on the MMTV promoter, similarly to that of GR, while it did not affect the transcriptional activity of NF-κB. These results suggest that Set/TAF-Iβ may be a general repressor of steroid receptor transcriptional activity.
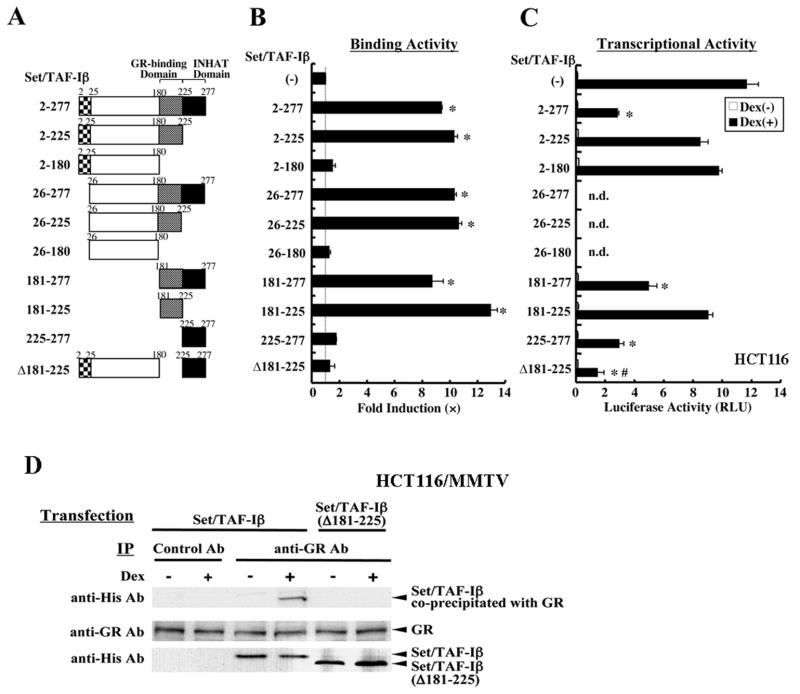
Set/TAF-Iβ interacts via its portion enclosed by amino acids 181-225 with the GR DBD
We next examined the interactions between Set/TAF-Iβ and the GR DBD in the reconstituted yeast two-hybrid assay to determine the portion of Set/TAF-Iβ necessary for physiologic association with this domain of the GR (Figure 2). Full-length Set/TAF-Iβ and its fragments containing amino acids 181 to 225 (GR-binding domain) including a short fragment of this portion, strongly interacted with the GR DBD, while fragments of Set/TAF-Iβ that did not include this portion, such as Set/TAF-Iβ-(Δ181–225) and -(2–180), lost the interaction (Figure 2A and B). In this binding assay, we also found that the INHAT domain of Set/TAF-Iβ, which mediated repressive effect of Set/TAF-Iβ on transcription, was not necessary for the interaction with the GR.
Figure 2. The Set/TAF-Iβ region included between amino acids 181 and 225 (GR-binding domain) is responsible for binding to the GR, while the INHAT domain (amino acids 225–277) is necessary and sufficient for the suppression of GR-induced transcriptional activity.
A: Linearized Set/TAF-Iβ fragments employed.
The GR-binding (amino acids 181–225) and INHAT (amino acids 225–277) domains are shown in gray and black, respectively.
B: The Set/TAF-Iβ-(181–225) binds to the GR DBD in a yeast two-hybrid assay. EGY48 yeast cells were transformed with p8OP-LacZ, pLexA-GRα-(420–489) and the indicated Set/TAF-Iβ-fragment-expressing plasmids. Bars represent the mean ± S.E. of fold activation compared to the baseline (the value obtained in the absence of Set/TAF-Iβ expression). *: p<0.01 (unpaired two-tailed Student t test ), compared to the baseline.
C: The INHAT domain (amino acids 225–277) is sufficient for the suppression of GR-induced transcriptional activity in HCT116 cells.
HCT116 cells were transfected with the indicated Set/TAF-Iβ fragment-expressing plasmid together with pRShGRα, pMMTV-Luc and pSV40-β-Gal. Open and solid bars indicate the mean ± S.E. of luciferase activity in the absence or presence of 10−6 M dexamethasone (Dex). *: p<0.01(unpaired two-tailed Student t test), compared to baseline (value obtained in the absence of Set/TAF-Iβ expression). #: p<0.01 (unpaired two-tailed Student t test), compared to “2–277” in the presence of dexamethasone. n.d.: not determined.
D: Set-TAF-Iβ is associated through amino acids 181 to 225 with the GR in vivo. HCT116/MMTV cells were transfected with the wild type His-Set-TAF-Iβ-or His-Set-TAF-Iβ(Δ181–225)-expressing plasmid together with pRShGRα, treated with 10−6 M of dexamethasone, and GR- Set-TAF-Iβ complexes were precipitated with control or anti-GR antibody. His-Set-TAF-Iβ-, His-Set-TAF-Iβ(Δ181–225) and GR were visualized with anti-His and anti-GR antibodies. IP results are shown in the top panel, whereas expressed GR, His-Set-TAF-Iβ and His-Set-TAF-Iβ(Δ181–225) are demonstrated in the bottom 2 panels.
We next examined the effects of these Set/TAF-Iβ fragments on GR-induced transcriptional activity to address the functional relevance of the observed interaction between Set/TAF-Iβ and the GR (Figure 3C). Set/TAF-Iβ fragments, such as Set/TAF-Iβ-(2–180) and -(2–225), which did not contain the INHAT domain (amino acids 255–277), lost their suppressive effect on GR-induced transcriptional activity regardless of the presence of the GR-binding domain (amino acids 181–225). Conversely, short Set/TAF-Iβ fragments (181–277) and (225–277), which contained the INHAT domain, actively suppressed GR transcriptional activity. Set/TAF-Iβ-(Δ181–225) had a stronger suppressive effect on GR-induced transcriptional activity than the wild type Set/TAF-Iβ. These results indicate that the INHAT domain, which was previously shown to mediate INHAT activity [9], is necessary and sufficient for suppression of GR-induced transcriptional activity. The GR-binding domain of Set/TAF-Iβ is, hence, dispensable for this activity. Indeed, removal of the domain from Set/TAF-Iβ made the fragment a stronger suppressor of GR transcriptional activity.
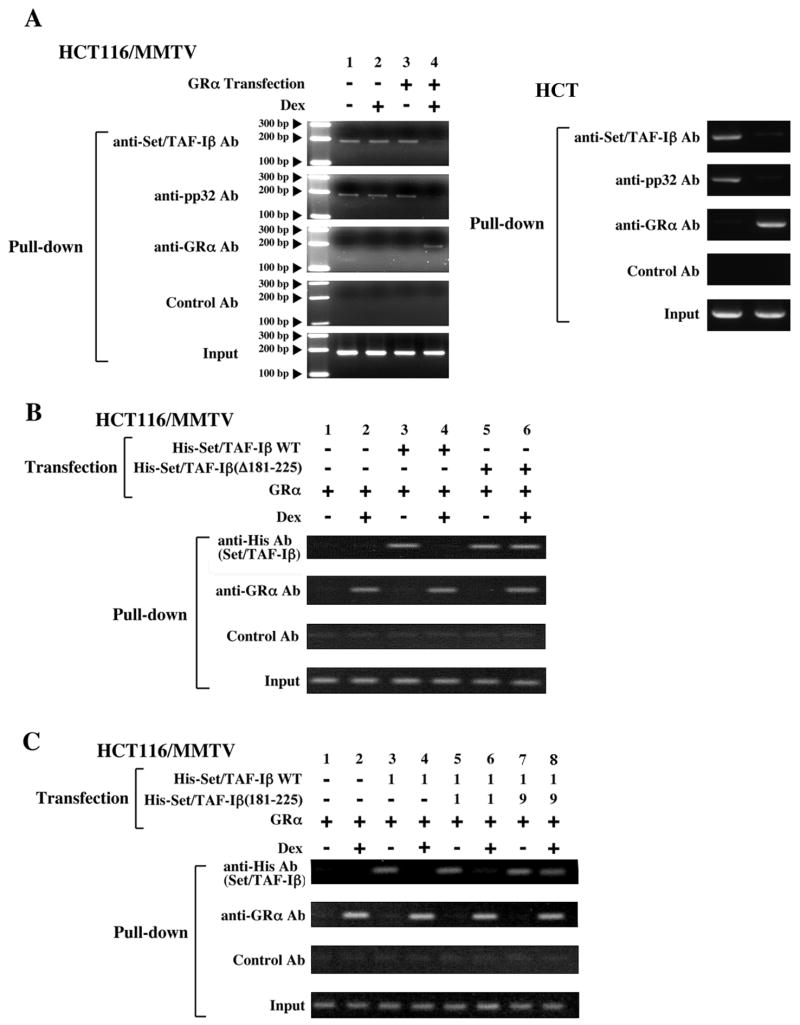
Figure 3. Binding of GR to GREs releases pre-existing INHAT complex from GREs via physical interaction between the GR and Set/TAF-Iβ.
A: Ligand-activated GR releases INHAT components Set/TAF-Iβ and pp32 from the MMTV GREs in HCT116/MMTV cells (left panels) and TAT GREs in HTC cells (right panels)
HCT116/MMTV cells transfected with pRShGRα or the control plasmid and HTC cells were treated with 10−6 M of dexamethasone (Dex). They were fixed with 1% formaldehyde and ChIP assays were performed by using anti-Set/TAF-Iβ, anti-pp32, anti -GRα or control antibodies.
B: Ligand-activated GR releases the wild type Set/TAF-Iβ but not the GR-binding domain-defective Set/TAF-Iβ-(Δ181–225) from the MMTV-GREs in HCT116/MMTV cells.
HCT116/MMTV cells were transfected with His-Set/TAF-Iβ-expressing plasmids and/or pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed by using anti-His, anti-GRαor control antibodies.
C: Overexpression of Set/TAF-Iβ-(181–225) attenuates GR-induced release of Set/TAF-Iβ from GREs in HCT116/MMTV cells. HCT116/MMTV cells were transfected with 0.5 μg/well of His-Set/TAF-Iβ-expressing plasmid (as “1”), pRShGRα and the indicated amounts (ratios) of the Set/TAF-Iβ-(Δ181–225)-expressing plasmid. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed using anti-His, anti-GRαor control antibodies.
We also examined interaction of GR and Set/TAF-Iβ in a co-immunoprecipitation assay (Figure 2D). HCT116 cells were transfected with His-Set/TAF-Iβ or His-Set/TAF-Iβ-(Δ181–225)-expressing plasmid together with the GR-expressing plasmid, and the protein complexes were precipitated with anti-GR antibody. Full length Set/TAF-Iβ was associated with GR in a dexamethasone-dependent fashion, while Set/TAF-Iβ-(Δ181–225) lost the interaction (Figure 2D, top panel). We observed co-precipitation of GR and Set/TAF-Iβ in the presence of the cross-linking agent hydroxylamine HCl, while such association was not found in its absence (data not shown), indicating that the physical interaction between GR and Set/TAF-Iβ is relatively weak. The Set/TAF-Iβ-related molecules were equally expressed throughout the experiment (Figure 2D, bottom panel). These results indicate that GR interacts with Set/TAF-Iβ through the latter’s GR-interacting domain in a ligand-dependent fashion in vivo.
GR releases Set/TAF-Iβ and pp32 from GREs in response to dexamethasone, while the GR-binding domain of Set/TAF-Iβ supports removal of Set/TAF-Iβ from GREs in chromatin immunoprecipitation (ChIP) assays
We examined the effect of GR on the association of INHAT to MMTV GREs by using the ChIP assay in HCT116/MMTV cells (Figure 3A, left panels). We employed a primer pair that detected the second and third GREs of the MMTV promoter [8,26]. The endogenous INHAT components, Set/TAF-Iβ and pp32, were constitutively co-precipitated with the MMTV GREs in the absence of GR transfection, possibly through their direct binding to histones that are associated with MMTV GREs (Figure 3A, left panels, top two panels, lanes 1, 2 and 3). Once GR was transfected, dexamethasone addition released these INHAT components from MMTV GREs (Figure 3A, left panels, top 3 panels, lane 4). We also performed the same ChIP assay in HTC cells, which express endogenous GR, Set/TAF-Iβ and pp32. The INHAT components Set/TAF-Iβ and pp32 were co-precipitated with TAT GREs in the absence of dexamethasone, while dexamethasone addition released them from the GREs. These results indicate that ligand-activated GR releases INHAT from GRE-associated histones.
To address whether physical association between Set/TAF-Iβ and GR is required for the removal of Set/TAF-Iβ from the MMTV GREs/histones, we transfected His-tagged wild type Set/TAF-Iβ or Set/TAF-Iβ-(Δ181–225) into HCT116/MMTV cells and performed ChIP assays (Figure 3B). GR-expressing plasmid was co-transfected throughout the experiment, as these cells did not express functional GR. Wild type Set/TAF-Iβ was co-precipitated with MMTV GREs in the absence of dexamethasone, while it lost this interaction in the presence of the steroid (Figure 3B, top panel, lanes 3 and 4), consistent with the results obtained in Figure 3A. In contrast, while Set/TAF-Iβ-(Δ181–225) was also co-precipitated with MMTV GREs in the absence of dexamethasone, it remained continuously associated with MMTV GREs in the presence of dexamethasone (Figure 3B, top panel, lanes 5 and 6). These results indicate that the physical interaction between Set/TAF-Iβ and GR through the GR-binding domain is necessary for the ligand-activated GR to release Set/TAF-Iβ from GRE-associated histones. GR was able to bind GREs in the presence of Set/TAF-Iβ-(Δ181–225) (Figure 3B, second top panel, lanes 5 and 6), consistent with previous findings that Set/TAF-Iβ binds histone tails but not DNA [9].
We further examined co-precipitation of the wild type Set/TAF-Iβ with the MMTV GREs in ChIP assays by over-expressing the GR-binding domain of Set/TAF-Iβ in HCT116/MMTV cells (Figure 3C). Increased expression of this fragment attenuated dexamethasone-induced release of Set/TAF-Iβ from MMTV GREs (Figure 3C, top panel, lanes 4, 6 and 8). These results further support our conclusion that GR releases the promoter-bound Set/TAF-Iβ through physical interaction between its DBD and the GR-binding domain of Set/TAF-Iβ.
Set/TAF-Iβ represses HAT-induced enhancement of GR transcriptional activity
The INHAT complex antagonizes histone acetylation caused by several HAT coactivators, such as CBP, p300 and p/CAF [9]. Thus, we examined Set/TAF-Iβ effects on HAT-induced enhancement of GR transcriptional activity in HCT116 cells (Figure 4A). Indeed, CBP, GRIP1 and p/CAF significantly enhanced GR-induced transactivation of the MMTV promoter in a dexamethasone-dependent fashion, while Set/TAF-Iβ suppressed the transcriptional enhancement of all HAT coactivators tested.
Figure 4. Set/TAF-Iβ and GRIP1 act antagonistically on GR-induced transcriptional activity.
A: Set/TAF-Iβ attenuates HAT coactivator-induced enhancement of GR-stimulated transcriptional activity in HCT116 cells.
HCT116 cells were transfected with pRShGRα together with indicated coactivator-expressing plasmid, and pMMTV-Luc in the absence or presence of Set/TAF-Iβ-expressing plasmid. Bars represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6 M of dexamethasone (Dex). *: p<0.01 (unpaired two-tailed Student t test), obtained by comparing the two values indicated.
B: Ligand-activated GR releases MMTV promoter-bound Set/TAF-Iβ from GREs, while it attracts the HAT coactivator GRIP1 to GREs in HCT116/MMTV cells.
HCT116/MMTV cells were transfected with GRIP1-expressing plasmid, His-Set/TAF-Iβ-expressing plasmid and/or pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed by using anti-His, anti-GRα, anti-GRIP1 or control antibodies.
C: Set/TAF-Iβ-(Δ181–225), a mutant defective in the GR-binding domain, is constitutively associated with GREs but does not inhibit dexamethasone-induced attraction of GRIP1 to GREs.
HCT116/MMTV cells were transfected with His-Set/TAF-Iβ-expressing plasmid, GRIP1-expressing plasmid, and/or pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed by using anti-His, anti-GRIP1, and anti-GRα antibodies.
D: Endogenous Set/TAF-Iβ suppresses dexamethasone-induced acetylation of histone H3 (K14) associated with GREs.
HCT116/MMTV cells were transfected with control or Set/TAF-Iβ siRNA, together with pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed by using anti-acetylated H3 (K14) antibody. Bars indicate fold acetylation of histone H3 (K14) (left panel) and fold induction of the Set/TAF-Iβ mRNA expression compared to that of the baseline. *: p<0.01, (unpaired two-tailed Student t test), compared to baseline (“Set/TAF-Iβ siRNA (−)” in presence of dexamethasone).
E: Set/TAF-Iβ-(Δ181–225), a mutant defective in the GR-binding domain, suppresses GRIP1-induced enhancement of dexamethasone-stimulated acetylation of histone H3 (K14) more strongly than the wild type Set/TAF-Iβ.
HCT116/MMTV cells were transfected with His-Set/TAF-Iβ wild type or (Δ181–225)-expressing plasmid and/or GRIP1-expressing plasmid, together with pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed using anti-acetylated H3 (K14) antibody. Bars indicate fold acetylation of histone H3 (K14) above the baseline. *: p<0.01 (unpaired two-tailed Student t test), compared to the baseline (the value obtained in the absence of plasmid transfection) or obtained by comparing the two values indicated.
We also examined the co-precipitation of Set/TAF-Iβ and GRIP1 with the MMTV promoter in the absence and presence of dexamethasone in HCT116/MMTV cells (Figure 4B). Set/TAF-Iβ was co-precipitated with MMTV GREs in the absence of dexamethasone, while it was released from these elements by addition of the steroid (Figure 4B, top panel, lanes 7 and 8). In contrast, GRIP1 was attracted to GREs only in the presence of dexamethasone (Figure 4B, third top panel, lanes 4 and 8). These results indicate that ligand-activated GR facilitates release of the INHAT complexes from and attraction of the HAT complexes to MMTV GREs in response to ligand. The GR DBD appears to mediate the release of the INHAT complexes by physically interacting with the GR-interacting domain of Set/TAF-Iβ, while the GR transactivation domains attract the HAT coactivators.
Since Set/TAF-Iβ-(Δ181–225) remained associated with MMTV GREs even in the presence of dexamethasone, we examined whether this Set/TAF-Iβ mutant influences GR-induced attraction of GRIP1 to the MMTV promoter in ChIP assays (Figure 4C). Wild type Set/TAF-Iβ was released from MMTV GREs by dexamethasone addition while Set/TAF-Iβ-(Δ181–225) remained associated with the GREs in its presence, consistent with the previous results (Figure 4C, top panel, lanes 2 and 4). GRIP1 was attracted to MMTV GREs by dexamethasone addition both in the presence of wild type Set/TAF-Iβ and Set/TAF-Iβ-(Δ181–225) (Figure 4C, 2nd top panel, lanes 2 and 4). These results indicate that Set/TAF-Iβ-(Δ181–225) and GRIP1 do not compete with each other for associating with MMTV GREs, consistent with previous findings that the former physically binds to GRE-associated histones, while the latter interacts with dexamethasone-activated and GRE-bound GR [6,9].
We examined the influence of Set/TAF-Iβ on the acetylation of histone H3 (Lysine 14), which is a known target of the INHAT [9], by using the siRNA for Set/TAF-Iβ in HCT116/MMTV cells (Figure 4D). Transfection of Set/TAF-Iβ siRNA strongly suppressed mRNA levels of endogenous Set/TAF-Iβ and enhanced dexamethasone-induced acetylation of histone H3, indicating that endogenous Set/TAF-Iβ protects histone H3 from acetylation induced by dexamethasone-stimulated GR. Therefore, we further examined competition of Set/TAF-Iβ and GRIP1 for the acetylation of histone H3 (K14) in HCT116/MMTV cells (Figure 4E). Overexpression of wild type Set/TAF-Iβ or Set/TAF-Iβ-(Δ181–225) suppressed acetylation of histone H3 associated with MMTV GREs. The GR-binding defective mutant suppressed the acetylation more strongly than the wild type Set/TAF-Iβ. Overexpression of GRIP1 enhanced histone H3 acetylation, while the wild type Set/TAF-Iβ and Set/TAF-Iβ-(Δ181–225) antagonized GRIP1-mediated enhancement of histone H3 acetylation. Again, Set/TAF-Iβ-(Δ181–225) antagonized GRIP1-induced acetylation more potently than the wild type Set/TAF-Iβ. Taken together, these results indicate that INHAT and the HAT complexes compete with each other for acetylation of histones, with INHAT suppressing GR-induced transcriptional activity. Binding of GR to Set/TAF-Iβ appears to facilitate removal of INHAT from GRE-associated histones, partially attenuating the inhibitory effect of INHAT on GR-induced transcriptional activity.
Set-Can fusion protein represses GR-induced transcriptional activity and ligand-activated GR fails to release this mutant protein from GREs
Set/TAF-Iβ forming a fusion protein with Can is a known oncoprotein in patients with acute undifferentiated leukemia [13,24]. Since these patients frequently demonstrate resistance to multiple chemotherapeutic compounds, including glucocorticoids [13,34,35], we hypothesized that the Set-Can fusion protein might cause glucocorticoid insensitivity in leukemic cells from patients by disrupting the physiologic regulation of GR-induced transcriptional activity by Set/TAF-Iβ. In reporter assays, Set-Can suppressed GR-induced transcriptional activity of the MMTV promoter in HCT116 cells more potently than Set/TAF-Iβ (Figure 5A).
Figure 5. The pathologic Set-Can fusion oncoprotein represses dexamethasone-stimulated transcriptional activity of the MMTV promoter by antagonizing GRIP1-induced acetylation of histone H3.

A: Set-Can suppresses GR-induced transcriptional activity in HCT116 cells.
HCT116 cells were transfected with TAF-Iβor Set/Can-expressing plasmid together with pRShGRα, pMMTV-Luc and pSV-40-β-Gal. Bars represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6 M of dexamethasone (Dex). *: p<0.01 (unpaired two-tailed Student t test), compared to the baseline (“Control” in the presence of dexamethasone), or obtained by comparing the two values indicated.
B: Set-Can is not physically associated with the GR in vivo.
HCT116/MMTV cells were transfected with the wild type His-Set-TAF-Iβ-or His-Set-Can-expressing plasmid together with pRShGRα and were treated with 10−6 M of dexamethasone. The protein complexes were precipitated with control or anti-GR antibody and His-Set-TAF-Iβ-, His-Set-Can and GR were visualized with anti-His and anti-GR antibodies. IP results are shown in the left panel, while expressed His-Set-TAF-Iβ and His-Set-Can were demonstrated in the right panel. GR expressed was also shown in the bottom panel.
C: Set-Can represses GRIP1-induced enhancement of GR transcriptional activity in HCT116/MMTV cells.
HCT116/MMTV cells were transfected with pRShGRα together with pMMTV-Luc and pSV40-β-Gal in the absence or presence of GRIP1- and/or Set/TAF-Iβ-expressing plasmids. Bars represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6 M of dexamethasone (Dex). Numbers on the bars indicate mean ± S.E values of GRIP1-induced fold enhancement of GR transcriptional activity in the absence and presence of Set-Can.
D: Set-Can dose-dependently represses GRIP1-induced enhancement of GR transcriptional activity in HCT116/MMTV cells.
HCT116/MMTV cells were transfected with the indicated amounts of Set/TAF-Iβ-expressing plasmid, together with 0.5 μg/well of the GRIP1-expressing plasmid, pRShGRα and pSV40-β-Gal, and pMMTV-Luc. Bars represent mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6 M of dexamethasone (Dex). *: p<0.01 (ANOVA, followed by Bonferroni correction), compared to the baseline (the value obtained in the presence of GRIP1 transfection and absence of Set-Can transfection).
E: Set-Can is constitutively associated with GREs in HCT116/MMTV cells.
HCT116/MMTV cells were transfected with His-Set/TAF-Iβ-or His-Set-Can-expressing plasmid, and pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde and ChIP assays were performed by using anti-His, anti-GRα or control antibodies.
F: Set-Can is associated with GREs both in the absence and presence of dexamethasone.
HCT116/MMTV cells were transfected with His-Set-Can-expressing plasmid and GRIP1-expressing plasmid, together with pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde, and ChIP assays were performed by using anti-His, anti-GRIP1 or anti-GRα antibodies.
G: Set-Can suppresses GRIP1-induced enhancement of dexamethasone-stimulated acetylation of histone H3 (K14) more strongly than the wild type Set/TAF-Iβ.
HCT116/MMTV cells were transfected with 0.5 μg/well of His-Set/TAF-Iβ or His-Set-Can-expressing plasmid and/or 0.5 μg/well of GRIP1-expressing plasmid, together with 0.5 μg/well of pRShGRα. The cells were treated with 10−6 M of dexamethasone (Dex), fixed with 1% formaldehyde, and ChIP assays were performed by using anti-acetylated H3 (K14) antibody. Bars indicate fold acetylation of histone H3 (K14) above the baseline. *: p<0.01 (unpaired two tailed Student t test), compared to the baseline (the value obtained in the absence of plasmid transfection), or obtained by comparing the two values indicated.
We also examined the interaction of Set-Can with GR in the co-immunoprecipitation assay (Figure 5B). HCT116/MMTV cells were transfected with His-Set/TAF-Iβ-or His-Set-Can-expressing plasmid together with the GR-expressing plasmid. The protein complexes were precipitated with anti-GR or control antibody and associated Set/TAF-Iβ and Set-Can was detected with anti-His antibody. Set/TAF-Iβ was co-precipitated with GR in the presence of dexamethasone, while Set-Can did not interact with the GR regardless of whether dexamethasone was present (Figure 5B, left panel). Amounts of Set/TAF-Iβ expressed were greater than those of Set-Can from the same amounts of plasmid added (Figure 5B, right panel), thus Set-Can appears to be a more potent inhibitor of GR transcriptional activity than Set/TAF-Iβ based on the results in Figure 5A.
We further examined the effect of Set-Can on GRIP1-induced enhancement of GR transcriptional activity in HCT116/MMTV cells (Figure 5C and D). GRIP1 enhanced GR-induced transcriptional activity of the MMTV promoter by 4.5 ± 0.25-fold and co-transfection of Set-Can reduced the GRIP1-induced enhancement by 2.8 ± 0.34-fold (p<0.05, n=3) (Figure 5C). We also examined increasing expression of Set-Can on GRIP1-induced enhancement of GR transcriptional activity in HCT116/MMTV cells (Figure 5D). Set-Can dose-dependently attenuated GRIP1 enhancement of GR-induced transcriptional activity. These results indicate that Set-Can interferes with GRIP1 preventing it from enhancing GR transcriptional activity.
We also examined the association of Set-Can and GRIP1 with MMTV GREs in ChIP assays (Figure 5E). Set-Can was constitutively co-precipitated with MMTV GREs not only in the absence, but also in the presence of dexamethasone, in contrast to Set/TAF-Iβ, which was released from GREs in the presence of the steroid (Figure 5E, top panel, lanes 3, 4, 5 and 6). GRIP1 was attracted to MMTV GREs in the presence of dexamethasone even though Set-Can was also precipitated with MMTV GREs (Figure 5F top two panels, lanes 2 and 4).
We examined dexamethasone-induced acetylation of histone H3 (K14) associated with MMTV GREs in the presence or absence of Set-Can and GRIP1 overexpression (Figure 5G). Overexpression of Set-Can suppressed dexamethasone-stimulated acetylation of histone H3 and suppressed GRIP1-induced enhancement of dexamethasone-stimulated histone acetylation much more potently than the wild type Set/TAF-Iβ. These results indicate that the pathologic fusion protein Set-Can suppresses GR-induced transcriptional activity by antagonizing HAT-induced acetylation of histones, in continuous association with GRE-bound histones due to its inability to communicate with ligand-activated and GRE-bound GR.
DISCUSSION
The INHAT complex suppresses the transcriptional activity induced by several transcription factors and nuclear receptors [21–23,32]. It does so at the baseline state by binding to lysine residues of the N-terminal tails of histones and by protecting them from acetylation induced by several HAT-coactivator complexes attracted to GRE-containing promoters by ligand-activated GR [32,36,37]. Set/TAF-Iβ, a component of the INHAT complex, physically interacts with these trans-acting factors mainly at their DBD [21,22,32]. In this study, we demonstrated that Set/TAF-Iβ, a major component of the INHAT complex, interacted with the GR DBD and suppressed GR-induced transcriptional activity of the chromatin-integrated MMTV promoter in HCT116 cells. GR interacted with Set/TAF-Iβ at a GR-binding domain (amino acids 181–225), adjacent N-terminally to the INHAT domain, consistent with a previous report that showed that the middle portion (amino acids 113–225) of Set/TAF-Iβ interacted with ERα in vitro [32]. The GR-binding domain of Set/TAF-Iβ was not necessary for suppression of GR-induced transcriptional activity but was essential for ligand-activated GR-mediated release of the INHAT complex from MMTV and TAT GREs (Figure 6A). A previous report indicated that Set/TAF-Iβ facilitated the interaction between ERα and estrogen response elements (EREs) in gel-shift assays [32], however, whether other transcription factors interact physically with Set/TAF-Iβ and whether such interactions play a role in their actions has not been elucidated as yet. In this report, we demonstrated that direct binding of GR to Set/TAF-Iβ was required for GR to release the INHAT complex from GRE-associated histones and to facilitate subsequent HAT-induced acetylation of histones. Physical interaction between GR and Set/TAF-Iβ appeared relatively weak, as it was observed only in the presence of a cross-linking compound, suggesting that physical interaction of GR to Set/TAF-Iβ may be transient and subtle on the target promoters. It was also reported that the INHAT complex was required for the in vitro transcription of a chromatin template, cooperating with ancillary factors like the ATP-dependent chromatin assembly factor [17]. Thus, Set/TAF-Iβ appears to be a dynamic transcriptional regulator of GR-induced transcriptional activity.
Figure 6. Activated GR releases INHAT from GREs through physical interaction with Set/TAF-Iβ, permitting HAT-induced acetylation of histone tails and hence gene transcription, while Set-Can suppresses GR transcriptional activity by being continuously associated with GREs and preventing GR coactivator-driven acetylation.

A: INHAT is associated with a GRE-containing promoter at the baseline state, binding histones and protecting them from transcription factor/nuclear receptor coactivator HAT-mediated acetylation. Once ligand becomes available, activated GR induces dissociation of INHAT from histones through physical interaction with Set/TAF-Iβ, making these histones susceptible to HAT-induced acetylation.
B: Pathologic Set-Can fusion protein remains associated with GREs in the presence of glucocorticoids, suppressing GR-induced transcriptional activity by preventing HAT-induced histone acetylation.
INHAT: inhibitor of the histone acetyltransferases, HAT: histone acetyltransferase, GR: glucocorticoid receptor, GREs: glucocorticoid response elements.
Set/TAF-Iβ forming a fusion with Can was originally discovered and cloned as an oncogene in patients with acute undifferentiated leukemia [13,24]. The role of the Set-Can fusion oncoprotein in the development of leukemia remains unknown, but Set/TAF-Iβ is highly expressed in proliferating cells and its overexpression induces differentiation in monocytic U937T cells [15]. We found that this fusion protein was not released from histones associated with GREs in response to ligand-activated GR and strongly suppressed GR-induced transcriptional activity by inhibiting HAT-induced histone acetylation. These results suggest that the Set-Can-mediated loss of regulation of histone acetylation is a potential mechanism of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation (Figure 6B).
The prognosis of patients with acute undifferentiated leukemia with Set-Can translocation is usually poor, with these patients frequently demonstrating resistance to chemotherapy regimens that include high doses of glucocorticoids [13,34,35]. This indicates that Set-Can might cause insensitivity of leukemic cells to glucocorticoids. It would be thus interesting to examine the expression levels and/or function of Set/TAF-Iβ in other types of leukemias and to correlate these values with the patients’ prognosis, responsiveness to glucocorticoid therapy and course of the disease.
Acknowledgments
This study was funded by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD. We appreciate the help of Drs. D. Chakravarti and L.K. Nieman, who critically read the manuscript and gave useful suggestions. We thank Drs. D. Chakravarti, R.M. Evans, G. Grosveld, G.L. Hager, K. Nagata, S.S. Simons Jr., M.R. Stallcup and N. Warriar for the plasmids provided and Mr. K. Zachman for superb technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chrousos GP. The hypothalamic-pituitary-adrenal axis and immune-mediated inflammation. N Engl J Med. 1995;332:1351–1362. doi: 10.1056/NEJM199505183322008. [DOI] [PubMed] [Google Scholar]
- 2.Chrousos GP. Glucocorticoid therapy. In: Felig P, Frohman LA, editors. Endocrinology & Metabolism. McGraw-Hill; New York: 2001. pp. 609–632. [Google Scholar]
- 3.Kino T, De Martino MU, Charmandari E, Mirani M, Chrousos GP. Tissue glucocorticoid resistance/hypersensitivity syndromes. J Steroid Biochem Mol Biol. 2003;85:457–467. doi: 10.1016/s0960-0760(03)00218-8. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11(Suppl 1):S45–55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 5.Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid receptors and associated diseases. Essays Biochem. 2004;40:137–155. doi: 10.1042/bse0400137. [DOI] [PubMed] [Google Scholar]
- 6.McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 7.Jones PL, Shi YB. N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. Curr Top Microbiol Immunol. 2003;274:237–268. doi: 10.1007/978-3-642-55747-7_9. [DOI] [PubMed] [Google Scholar]
- 8.Ichijo T, Voutetakis A, Cotrim AP, Bhattachryya N, Fujii M, Chrousos GP, Kino T. The Smad6-histone deacetylase 3 complex silences the transcriptional activity of the glucocorticoid receptor: potential clinical implications. J Biol Chem. 2005;280:42067–42077. doi: 10.1074/jbc.M509338200. [DOI] [PubMed] [Google Scholar]
- 9.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 10.Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Natl Acad Sci U S A. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti KG, Davis D, Bonten J, Buijs A, Grosveld G. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- 12.Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- 13.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito S, Miyaji-Yamaguchi M, Shimoyama T, Nagata K. Functional domains of template-activating factor-I as a protein phosphatase 2A inhibitor. Biochem Biophys Res Commun. 1999;259:471–475. doi: 10.1006/bbrc.1999.0790. [DOI] [PubMed] [Google Scholar]
- 15.Carlson SG, Eng E, Kim EG, Perlman EJ, Copeland TD, Ballermann BJ. Expression of SET, an inhibitor of protein phosphatase 2A, in renal development and Wilms’ tumor. J Am Soc Nephrol. 1998;9:1873–1880. doi: 10.1681/ASN.V9101873. [DOI] [PubMed] [Google Scholar]
- 16.Miyaji-Yamaguchi M, Okuwaki M, Nagata K. Coiled-coil structure-mediated dimerization of template activating factor-I is critical for its chromatin remodeling activity. J Mol Biol. 1999;290:547–557. doi: 10.1006/jmbi.1999.2898. [DOI] [PubMed] [Google Scholar]
- 17.Gamble MJ, Erdjument-Bromage H, Tempst P, Freedman LP, Fisher RP. The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol Cell Biol. 2005;25:797–807. doi: 10.1128/MCB.25.2.797-807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto K, Nagata K, Ui M, Hanaoka F. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J Biol Chem. 1993;268:10582–10587. [PubMed] [Google Scholar]
- 19.Cervoni N, Detich N, Seo SB, Chakravarti D, Szyf M. The oncoprotein Set/TAF-1β, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277:25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- 20.Okuwaki M, Nagata K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J Biol Chem. 1998;273:34511–34518. doi: 10.1074/jbc.273.51.34511. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Muto S, Miyamoto S, Aizawa K, Horikoshi M, Nagai R. Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J Biol Chem. 2003;278:28758–28764. doi: 10.1074/jbc.M302228200. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol. 2003;23:8528–8541. doi: 10.1128/MCB.23.23.8528-8541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macfarlan T, Parker JB, Nagata K, Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Iβ and inhibits histone acetylation to repress transcription. Mol Endocrinol. 2006;20:335–347. doi: 10.1210/me.2005-0248. [DOI] [PubMed] [Google Scholar]
- 24.von Lindern M, Breems D, van Baal S, Adriaansen H, Grosveld G. Characterization of the translocation breakpoint sequences of two DEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and a SET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosomes Cancer. 1992;5:227–234. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- 25.Mirani M, Elenkov I, Volpi S, Hiroi N, Chrousos GP, Kino T. HIV-1 protein Vpr suppresses IL-12 production from human monocytes by enhancing glucocorticoid action: potential implications of Vpr coactivator activity for the innate and cellular immunity deficits observed in HIV-1 infection. J Immunol. 2002;169:6361–6368. doi: 10.4049/jimmunol.169.11.6361. [DOI] [PubMed] [Google Scholar]
- 26.Bresnick EH, John S, Berard DS, LeFebvre P, Hager GL. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci U S A. 1990;87:3977–3981. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kino T, Gragerov A, Valentin A, Tsopanomihalou M, Ilyina-Gragerova G, Erwin-Cohen R, Chrousos GP, Pavlakis GN. Vpr protein of human immunodeficiency virus type 1 binds to 14-3-3 proteins and facilitates complex formation with Cdc25C: implications for cell cycle arrest. J Virol. 2005;79:2780–2787. doi: 10.1128/JVI.79.5.2780-2787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kino T, Tiulpakov A, Ichijo T, Chheng L, Kozasa T, Chrousos GP. G protein β interacts with the glucocorticoid receptor and suppresses its transcriptional activity in the nucleus. J Cell Biol. 2005;169:885–896. doi: 10.1083/jcb.200409150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kino T, Gragerov A, Kopp JB, Stauber RH, Pavlakis GN, Chrousos GP. The HIV-1 virion-associated protein vpr is a coactivator of the human glucocorticoid receptor. J Exp Med. 1999;189:51–62. doi: 10.1084/jem.189.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charmandari E, Raji A, Kino T, Ichijo T, Tiulpakov A, Zachman K, Chrousos GP. A novel point mutation in the ligand-binding domain (LBD) of the human glucocorticoid receptor (hGR) causing generalized glucocorticoid resistance: The importance of C-terminus of hGR LBD in conferring transactivational activity. J Clin Endocrinol Metab. 2005;90:3696–3705. doi: 10.1210/jc.2004-1920. [DOI] [PubMed] [Google Scholar]
- 31.De Martino MU, Bhattachryya N, Alesci S, Ichijo T, Chrousos GP, Kino T. The glucocorticoid receptor and the orphan nuclear receptor chicken ovalbumin upstream promoter-transcription factor II interact with and mutually affect each other’s transcriptional activities: implications for intermediary metabolism. Mol Endocrinol. 2004;18:820–833. doi: 10.1210/me.2003-0341. [DOI] [PubMed] [Google Scholar]
- 32.Loven MA, Muster N, Yates JR, Nardulli AM. A novel estrogen receptor α-associated protein, template-activating factor Iβ, inhibits acetylation and transactivation. Mol Endocrinol. 2003;17:67–78. doi: 10.1210/me.2002-0280. [DOI] [PubMed] [Google Scholar]
- 33.Jantzen HM, Strahle U, Gloss B, Stewart F, Schmid W, Boshart M, Miksicek R, Schutz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987;49:29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- 34.Griggs JJ, Henley SE, Rowe JM. Treatment of refractory undifferentiated acute myelogenous leukemia with all-trans-retinoic acid. Am J Hematol. 1994;45:177–180. doi: 10.1002/ajh.2830450215. [DOI] [PubMed] [Google Scholar]
- 35.Singhal S, Powles R, Treleaven J, Kulkarni S, Horton C, Mehta J. Long-term outcome of adult acute leukemia patients who are alive and well two years after allogeneic bone marrow transplantation from an HLA-identical sibling. Leuk Lymphoma. 1999;34:287–294. doi: 10.3109/10428199909050953. [DOI] [PubMed] [Google Scholar]
- 36.Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- 37.Kutney SN, Hong R, Macfarlan T, Chakravarti D. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Iβ in integrating chromatin hypoacetylation and transcriptional repression. J Biol Chem. 2004;279:30850–30855. doi: 10.1074/jbc.M404969200. [DOI] [PubMed] [Google Scholar]