Abstract
The tetraspanin CD81 is ubiquitously expressed and associated with CD19 on B lymphocytes and with CD4 and CD8 on T lymphocytes. Analysis of mice with disrupted CD81 gene reveals normal T cells but a distinct abnormality in B cells consisting of decreased expression of CD19 and severe reduction in peritoneal B-1 cells. CD81-deficient B cells responded normally to surface IgM crosslinking, but had severely impaired calcium influx following CD19 engagement. CD81-deficient mice had increased serum IgM and IgA and an exaggerated antibody response to the type II T independent antigen TNP-Ficoll. These results suggest that CD81 is important for CD19 signaling and B cell function.
CD81 is a ubiquitously expressed cell surface protein that belongs to the transmembrane-4 superfamily (TM4SF) (1, 2). TM4SF proteins are characterized by the presence of four conserved hydrophobic membrane spanning domains and consist to date of >15 members that include CD9, CD37, CD53, CD81, and CD82 (3). Little is known about the function of TM4SF proteins.
CD81 is a component of the CD19/CD21/Leu-13/CD81 signal transducing complex in B lymphocytes (4). CD81 associates directly with Leu-13 and CD19 and indirectly, via CD19, with CD21 (5–7). This complex plays an important costimulatory role in B cell activation. Crosslinking of CD19 to sIgM lowers the threshold required for activation through the B cell antigen receptor (BCR) (8). The CD21 component of the CD19/CD21 complex is a receptor for iC3b, C3dg, and C3d fragments of the third component of complement (9). Like CD19, it plays a important role in amplifying antibody responses. C3d cross-linked with antigen functions as a molecular adjuvant in the development of humoral immune responses (10). Furthermore, CD19 and CD21/CD35-deficient mice have impaired antibody responses to thymus dependent (TD) antigens, reduced germinal center formation, and impaired affinity maturation of serum antibodies (11–15).
CD81 and CD82 are associated with CD4 and CD8 molecules on the T cell surface (16, 17). Addition of anti-CD81 mAb to fetal thymus organ culture blocked the progression of immature thymocytes from double negative (CD4−CD8−) to the double positive (CD4+CD8+) stage (18). Single positive TCRαβ cells were absent in these cultures, while development of TCRγδ cells proceeded normally. These results suggested that interaction between immature thymocytes and stromal cells expressing CD81 is required to induce early events associated with the development of TCRαβ lymphocytes.
CD81 and at least three other TM4SF members (CD37, CD53, and CD82) coprecipitate with major histocompatibility complex class II molecules from human B cells and may participate in signal transduction via major histocompatibility complex class II molecules (19–21). CD81 and other TM4SF proteins are also associated with integrins on the cell surface. CD81 coprecipitates with α3β1, α4β1, α6β1, α4β2, and αEβ7, but not with α2β1, α5β7, and αLβ2 integrins (22–24). Integrins and TM4SF proteins both regulate cell motility and adhesion (25–28).
To understand the role of CD81 in lymphocyte development and function, we have generated mice with disrupted CD81 gene. Analysis of these mice indicate that CD81 is not important for T cell development, but plays an important role in CD19 expression and signaling and in B cell development and function.
MATERIALS AND METHODS
Generation of CD81-Deficient Mice.
DNA encoding the murine CD81 gene was isolated from a Lambda FIXII library made from the 129Sv mouse strain (Stratagene) using a human CD81 cDNA probe kindly provided by Shoshana Levy (Stanford University, Stanford, CA). DNA from isolated phages was purified and subjected to high resolution restriction mapping by partial digestion and Southern blot analysis. The targeting construct was assembled using the 6.5-kb XhoI/SalI and the 3.5-kb BamHI/KpnI fragments in the pPNT targeting vector (kindly provided by Richard Mulligan, Children’s Hospital, Boston). This vector contains the neomycin gene (neo) for positive selection of the transfected embryonic stem (ES) cells and a copy of the thymidine kinase gene (TK) for the negative selection of spurious transformants that contain randomly integrated constructs. The construct (20 μg) was then linearized by digestion at the unique NotI site in pPNT and used to transfect by electroporation 2 × 107 of ES cells (J1) obtained from R. Jaenisch (Massachusetts Institue of Technology, Cambridge). Transfected ES cells were selected by growing them in medium containing 0.4 mg/ml of G418 and 10 μg/ml of gancyclovir. Of 118 clones analyzed, 1 clone was identified to contain both a normal and a disrupted allele. The targeted ES clone was injected into 3.5-day-old C57BL/6 blastocysts, which were then transferred into Swiss foster mothers. Chimeric males were crossed with C57BL/6 females. Tail DNAs of agouti offsprings were analyzed by Southern blot. CD81 heterozygous (+/−) mice from the F1 generation were used to obtain CD81 homozygous (−/−) mice by brother-sister mating. F2 offsprings from these crosses were genotyped by Southern blot analysis to identify homozygotes.
Antibodies and Flow Cytometry Analysis.
All antibodies and streptavidin-Cy-Chrome used in this study were purchased from PharMingen unless otherwise specified. Expression of CD19 on B cells was determined by staining with anti-CD19 mAb (1D3, rat IgG2a) purchased from PharMingen and with anti-CD19 mAb (MB19-1, mouse IgA) obtained from Shihichi Sato (Durham, NC). Single cell suspensions from spleen, bone marrow, thymus, and peritoneal cavity were isolated on density gradient of Lympholyte-M (Cedarlane Laboratories). Peritoneal leukocytes were purified by i.p. lavage with ice-cold PBS. Cells were stained and analyzed on a FACSCalibur flow cytometer (Becton Dickinson) as previously described (29). Fluoresence-activated cell sorter analysis was performed on cells from at least six mice both at 4 and 8 weeks of age.
Serum Antibody Responses and Ig Levels.
Two-month-old mice were immunized i.v. with 100 μg of 2,4,6-trinitrophenyl (TNP)-keyhole limpet hemocyanin (KLH) conjugate (1:25) in PBS at day 0, boosted at day 21 and bled at days 0, 7, 14, 21, and 28. Mice were immunized i.p. with 10 μg lipopolysaccaride (LPS)-TNP (Sigma) in PBS or with 10 μg TNP-Ficoll (a gift of F. D. Finkelman, Bethesda) at day 0 and bled at days 0, 3, 7, and 14. Levels of antigen-specific antibody responses were analyzed by TNP-specific ELISA using 96-well plates coated with TNP-conjugated BSA at 10 μg/ml from PBS. Isotype-specific antibodies conjugated to biotin; strepavidin–peroxidase conjugates and recommendations for ELISA were obtained from PharMingen. Sera were diluted 1:1,000 and 1:5,000 and levels of Ig were determined.
B Cell Proliferation.
B cells were purified from single spleen cell suspension by depletion of T cells with anti-CD4, CD8, and Thy1.1 mAbs and magnetic beads BioMag conjugated with goat anti-rat IgG (PerSeptive Diagnostics, Cambridge, MA). B cells were cultured at 1 × 105 per well for 72 hr and activated by F(ab)′2 fragments of goat anti-IgM polyclonal antibodies (Rockland, Gilbertsville, PA), anti-CD40 mAb (PharMingen), LPS (Sigma), and anti-CD19 mAb MB19-1 at indicated concentrations. Proliferation was assessed by the incorporation of [3H]thymidine added (1 μCi per well; 1 Ci = 37 GBq) during the last 6 hr of culture.
Measurement of Intracellular Free Calcium ([Ca2+]i).
Analysis of [Ca2+]i was performed as described (30). Splenocytes were loaded with Fluo-3 by incubation of 1 × 107/ml cells in Hanks’ balanced saline solution (HBSS), containing Fluo-3/AM (2 μM) and pluronic F-127 (0.05%) for 40 min in a 37.5°C water bath. The cell suspension was diluted 1:5 with HBSS containing 1% fetal calf serum and incubated for 40 min. Fluo-3-loaded cells were washed and stained with B220-phycoerythrin (PE) antibody for 20 min at 4°C. The cells were then washed twice and diluted to 106/ml in 10 mM Hepes buffer (pH 7.4), containing 137 mM NaCl, 5 mM KCl, 1 mM Na2HPO4, 5 mM glucose, 1 mM CaCl2, 0.5 mM MgCl2, and 1 g/liter BSA. The cell suspension was placed in a water bath at 37°C for 15 min before flow cytometry. Analysis was performed by using FACSCalibur with cellquest software (Becton Dickinson). Forward light scatter and side light scatter were displayed on linear scale, whereas Fluo-3 and PE fluorescence were displayed on four-decade logarithmic scale. The “time” parameter was activated on a scale of 512 sec (500 msec per channel). B cell analysis was performed by gating on PE-positive cells. At 1 min after base-line measurement, F(ab′)2 goat anti-IgM or anti-CD19 mAb was added to the sample.
RESULTS AND DISCUSSION
Generation of CD81-Deficient Mice.
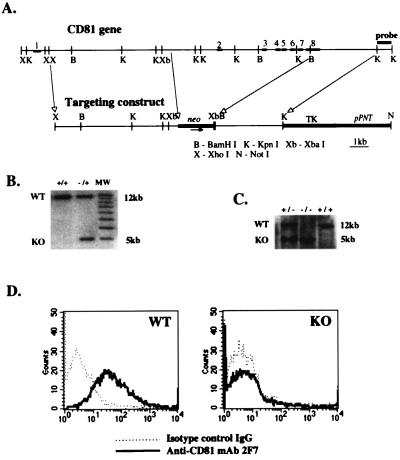
The structures of the CD81 gene and of the targeting construct are shown in Fig. 1A. Following homologous recombination, exons 2–8 of CD81, which include most of the coding region and their intervening introns, are replaced by a neomycin resistance gene. Homologous recombinant ES clones were identified by the presence of a novel restriction fragment of ≈5 kb derived from the targeted allele in addition to the ≈12-kb fragment derived from the wild-type allele (Fig. 1B) following digestion of genomic DNA with XbaI and probing with a KpnI–KpnI fragment derived from the region located immediately 3′ to the targeted locus. CD81 homozygous (−/−) mice were derived by standard techniques and identified by Southern blot analysis of tail DNA (Fig. 1C). Disruption of the CD81 gene was confirmed by demonstrating that thymocytes from CD81-deficient mice have no detectable CD81 surface expression by fluoresence-activated cell sorter analysis (Fig. 1D). CD81-deficient mice were raised in a germ free environment and do not display any differences from wild-type littermates in growth, weight, health, or reproductive potential.
Figure 1.
Generation of CD-81-deficient mice. (A) Structure of the CD81 gene (Upper) and of the targeting construct (Lower). Exons are represented by bold segments and are numbered. neo, neomycin resistance gene. (B) Southern blot analysis of DNA from ES cell clones. Genomic DNA was digested by XbaI and probed with the KpnI fragment shown in A. The wild-type (WT) allele is represented by the 5-kb band. (C) Southern blot analysis of DNA from tails of littermate mice of brother-sister mating of CD81 heterozygotes (+/−) mice. Analysis was performed as in B. (D) Flow cytometry analysis of surface expression of CD81 on thymocytes from WT and CD81-deficient (KO) mice. Cells were stained with biotinylated anti-CD81 hamster mAb 2F7 (——) or biotinylated hamster IgG as an isotype control (· · · ·) followed by streptavidin-PE.
Lymphocyte Development in CD81-Deficient Mice.
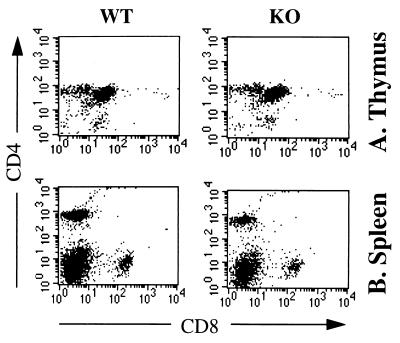
CD81-deficient mice have thymi and spleens of normal size and architecture. Detailed fluoresence-activated cell sorter analysis was performed on groups of at least six mice at ages 4 and 8 weeks with similar results. Analysis of mononuclear cells from thymus and spleen revealed normal cell numbers and normal expression of T cell markers as evidenced by expression of CD4, CD8, TCRαβ, TCRγδ, CD2, CD3, Thy-1, CD45R, CD5, CD25, and heat stable antigen (Fig. 2 and data not shown). These results indicate that CD81 expression is not necessary for T cell development. Moreover, T cell help to B cells in the antibody response to the TD antigen KLH-TNP was normal in CD81-deficient mice (see below), suggesting that T cell function was grossly normal. The previously reported inhibition of thymocyte development by anti-CD81 mAb (18) may be explained by a negative signal delivered by anti-CD81 mAb to the developing T cells directly or via stromal cells.
Figure 2.
Flow cytometry analysis of thymocytes (A) and spleen cells (B) from CD81-deficient mice. Single cell suspension from wild-type (WT) and CD81-deficient (KO) littermates were stained for CD4 [anti-CD4-fluorescein isothiocyanate (FITC)] vs. CD8 (anti-CD8-PE). Cells with forward and side light scatter properties of lymphocytes were analyzed by two-color flow cytometry.
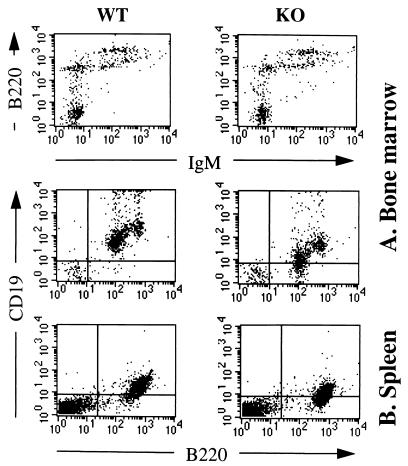
CD81-deficient mice had normal numbers of B cells in the bone marrow and spleen (data not shown). Fig. 3 shows that bone marrow from CD81-deficient mice had normal numbers of IgM−B220lo cells (pro-B cells), IgM−B220int cells (pre-B cells), IgM+B220int (immature B cells), and IgM+B220hi cells (mature B cells). Thus, all stages of B cell development in the bone marrow appear to be normal. Except for CD19, B cells cells had normal expression of surface markers including B220, sIgM (Fig. 3), sIgD, sIgG, CD5, CD21, CD22, CD23, CD40, CD43 (S7), and heat stable antigen (data not shown). CD19 expression was also reduced on spleen B cells (Fig. 3), whereas CD21 and CD40 expression was normal. The level of CD19 expression on bone marrow and spleen B cells was reduced by ≈2-fold as evidenced by ≈50% reduction in mean fluorescence intensity. CD81 is associated directly with CD19, but not with CD21 (6) and may facilitate the transport of CD19 to the cell surface and/or enhance its stability and/or retard its degradation. The apparent less intense staining for CD19 on spleen cells compared with mature bone marrow cells is due to the use of different fluorochromes, namely PE for bone marrow cells and FITC for spleen cells. When the same fluorochrome was used the intensities of CD19 expression on spleen and mature bone marrow cells were similar (data not shown).
Figure 3.
Flow cytometry of B lymphocytes in CD81-deficient mice (KO) and wild-type (WT) littermates. (A) Bone marrow cells were stained for IgM (anti-IgM-FITC) vs. B220 (anti-B220-PE); and for B220 (anti-B220-FITC) vs. CD19 (anti-CD19-PE). Cells with forward and side light scatter properties of lymphocytes were analyzed. (B) Spleen cells were stained for CD19 (anti-CD19-FITC) vs. B220 (anti-B220-PE).
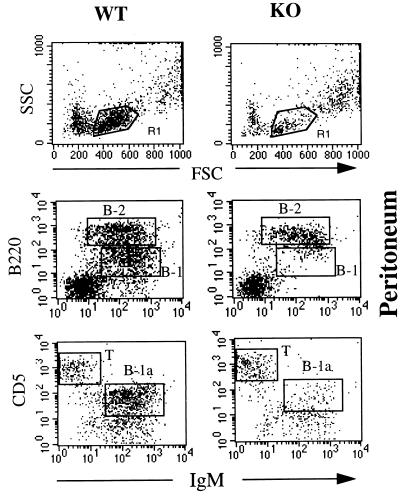
B-1 cells represent a major B cell population in the peritoneum (31). B-1 cells are distinguished from conventional B (B-2) cells by the expression of CD11b (Mac-1) and of intermediate levels of B220 (32, 33). B-1 cells are divided in two subpopulations on the basis of CD5 expression: B-1a (CD5+) and B-1b (CD5−) cells. There was a significant reduction in peritoneal cellularity in CD81-deficient mice as compared with wild-type mice, 0.9 ± 0.3 × 106 vs. 2.5 ± 0.44 × 106 cells per mouse, respectively (P < 0.05, n = 5 in each group). The number of granulocytes and monocytes was comparable in CD81-deficient and wild-type mice (data not shown). In contrast, the total number of lymphocytes was decreased by ≈80% in CD81-deficient mice as compared with controls (Fig. 4). There was severe reduction of IgM+B220int cells (Fig. 4) and of CD11 b+B220int cells (data not shown) that correspond to B-1 cells. A severe reduction in B-1a cells was evident by the dramatic decrease in IgM+CD5+cells (Fig. 4). In contrast, the numbers of peritoneal B-2 cells (IgMhiB220hi) and T cells (CD5hiIgM−) were normal. These results suggest that CD81 is critical for the development or self-renewal of peritoneal B-1 cells.
Figure 4.
Flow cytometry analysis of peritoneal cells. Cells were analyzed with forward (FSC) and side (SSC) light scatter. The gated (R1) lymphocyte population was stained for IgM (anti-IgM-FITC) vs. B20 (anti-b220-PE) and for IgM (anti-IgM-FITC) vs. CD5 (anti-CD5-PE). B-1 (B220intIgM+, B-2 (B220hiIgM+), B-1a (B220loIgM+), and T (CD5hiIgM−) cells are boxed.
Peritoneal B-1 cells are decreased in mice deficient in CD19, CD21, PKCβ, btk vav, IL-5, and IL-5R, all molecules that are important for B cell activation (11, 13, 15, 34–37). Conversely, peritoneal B-1 cells are increased in mice that lack SHP-1 and CD22, molecules that negatively regulate B cell activation (38–41). These findings support the notion that the generation of B-1 cells is dependent on their activation. The severe reduction in B-1 cells in CD81-deficient mice is in line with the observation that self-renewal of B-1 cell is dependent on the levels of CD19 expression (42, 43) and suggests that CD81 may play an important role in the cellular activation that is necessary for generating B-1 cells.
Impaired CD19 Signaling in B Cells from CD81-Deficient Mice.
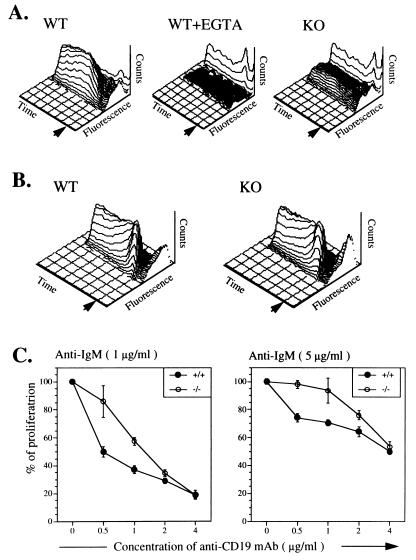
An important consequence of CD19 engagement is a rise in [Ca2+]i concentration (41, 44, 45). CD19 engagement on normal B cells with mAb MB19-1 resulted in a slow rise in [Ca2+]i, which lacked the initial rapid phase characteristic of calcium mobilization from intracellular stores and reached a maximum at 100–140 sec (Fig. 5A). This rise was virtually abolished by EGTA supporting that influx of extracellular calcium was its major contributor. B cells from CD81-deficient mice exhibited a very poor calcium influx in response to ligation of CD19 engagement with up to 40 μg/ml of anti-CD19 mAb. Even at this high concentration of anti-CD19 mAb, <20% of CD81-deficient B cells exhibited a rise in [Ca2+]i compared with >95% of wild-type B cells. Thus it is unlikely that this phenomenon is due to the 50% reduction in CD19 expression by CD81-deficient B cells. Formal demonstration that the impaired CD19-dependent calcium flux in CD81-deficient mice is not simply due to decreased CD19 expression will require expression of a CD19 transgene or of a mutant CD81 transgene that restore normal CD19 expression but not CD19-dependent calcium flux in CD81-deficient mice.
Figure 5.
Response of CD8-deficient B cells to CD19 ligation. (A) [Ca2+]i response to anti-CD19 mAb in B cells. Splenocytes from wild-type (WT) and CD81-deficient (KO) littermates were loaded with Fluo-3 and stained with anti-B220-PE. Analysis of [Ca2+]i was performed on gated B220-positive cells. The time scale represents 512 s. Cell counts (counts) are represented on the vertical axis. Arrow indicated the addition of anti-CD19 mAb MB19-1 at a concentration of 40 μg/ml. EGTA (10 mM) was added at time ). (B) [Ca2+]i response to anti-IgM in B cells. Arrow indicates the addition of F(ab′)2 goat anti-mouse IgM 1 μg/ml). (C) Modulation of B cell proliferation in response to anti-IgM by anti-CD19 mAbs. purified splenic B cells (1 × 105 cells per well) from WT (+/+) and CD81-deficient (−/−) littermates were stimulated with F(ab′)2 fragments of goat anti-mouse IgM (1 and 5 μg/ml) in the presence or absence of anti-CD19 mAb MB19-1 at the indicated concentrations. Results are expressed as percent of the proliferation of cell cultures stimulated with anti-IgM in the absence of anti-CD19 mAb. Each of the experiments shown is representative of four independent experiments.
In contrast to their impaired calcium mobilization in response to CD19 ligation, CD81-deficient B cells mobilized calcium normally in response to sIgM crosslinking (Fig. 5B). These results suggest that CD81 is critical for calcium influx following engagement of CD19 on B cells and raise the speculation that CD81 may function as a calcium-permeable cation channel regulated by CD19.
CD81 is characterized by the presence of four hydrophobic transmembrane spanning domains that are relatively rich in polar residues, including a highly conserved asparagine residue in transmembrane domain 1 and glutamic acid/glutamine residues in transmembrane domains 3 and 4 (3, 46). The same polar residues are conserved in transmembrane domains 1, 3, and 4 of the tetraspan B cell specific surface antigen CD20 (47), which unlike CD81 is not a TM4SF member, but which functions as a calcium-permeable cation channel (48). Cations permeating through the pore of an ion channel are thought to interact with the polar amino acids that line the channel pore (49). Mutational analysis of the polar amino acid residues in CD81 is in progress to determine their role in CD19-operated CD81-mediated calcium influx.
Crosslinking of CD19 independently of sIgM inhibits BCR signaling (44, 50, 51). B cells from CD81-deficient mice proliferated normally in response to F(ab′)2 anti-IgM (0.1, 1, and 10 μg/ml), as well as to anti-CD40 mAb and LPS (data not shown). Fig. 5 shows that CD81-deficient B cells were less sensitive than wild-type B cells to inhibition by anti-CD19 mAb used at 0.5 and 1 μg/ml. The decreased susceptibility of CD81-deficient B cells to inhibition by anti-CD19 mAb may be due to the 50% decrease in CD19 expression. Alternatively, impaired CD19-mediated calcium influx may interfere with the ability of CD19 to deliver an inhibitory signal in the absence of CD81.
Increased Serum IgM and IgA Levels and Enhanced Antibody Responses To Type II T Independent (TI) Antigen in CD81-Deficient Mice.
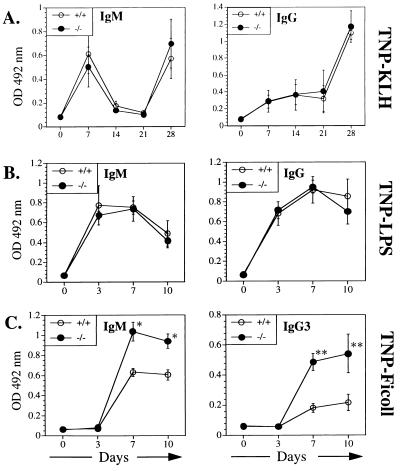
CD19- and CD21-deficient mice have markedly impaired TD humoral immune responses. This is thought result from inefficient trapping of complement fixing antigen complexes by the B cells in the absence of CD21 (12) and to reflect an important role of CD19 in initial B cell activation by TD antigens. Fig. 6 shows that both primary and secondary responses to the TD antigen KLH-TNP were normal in CD81-deficient mice. This suggests that CD81 is not critical for CD21-mediated internalization of protein antigen by B cells nor for the delivery of CD19-mediated costimulatory signals that are important in the response to TD antigens.
Figure 6.
Antibody responses of wild-type mice (+/+) and CD81-deficient (−/−) mice. (A) Response to the TD antigen TNP-KLH. Mice were immunized i.v. with 100 μg of TNP-KLH conjugate 1:25) in PBS at day 0 and boosted at day 21. (B) response to type I TI antigen TNP-LPS. Mice were immunized i.p. with 10 μg TNP-LPS in PBS at day 0. (C) Response to the type II TI antigen TNP-Ficoll. Mice were immunized i.p. with 10 μg TNP-Ficoll in PBS at day 0. Sera were diluted 1/1,000 and levels of antigen-specific antibody responses of the indicated isotypes were analyzed by TNP-specific ELISA. Mean values ± 2 SD obtained for the four mice per group are shown. ∗, P < 0.005; ∗∗, P < 0.01.
The antibody response of CD81-deficient mice to the type I TI antigen TNP-LPS was normal (Fig. 6). TNP-LPS delivers two activations signals to the B cell, one via the BCR and one by LPS. The normal response of CD81-deficient B cells to TNP-LPS is consistent with the normal proliferative response of these B cells to LPS and to BCR crosslinking.
CD81-deficient mice had a significantly enhanced antibody response to the type II TI antigen TNP-Ficoll in both IgM and IgG3 isotypes (Fig. 7). Type II TI antigens contain repetitive antigenic determinants that cause a high degree of sIgM crosslinking resulting in B cell activation and antibody production. The enhanced response of CD81-deficient B cells to crosslinking of the BCR in vivo by type II TI antigen, but not in vitro by anti-IgM, may be explained by the fact that CD81 may have an inhibitory effect on signaling pathways triggered by BCR crosslinking. Hyperresponsiveness of CD81-deficient mice to TNP-Ficoll may also relate to decreased CD19 expression because CD19-deficient mice also display augmented antibody responses to type II TI antigens (52) and even small changes in CD19 expression can have dramatic effects on B cell function (53).
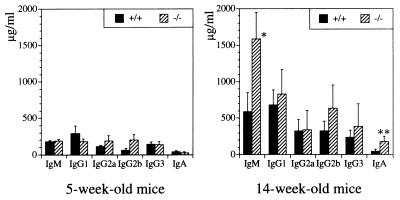
Figure 7.
Serum Ig levels in unimmunized wild-type (+/+) and CD81-deficient (−/−) mice. Mean values ± 2 SD obtained for six mice per group are shown. ∗, P < 0.005; ∗∗, P < 0.01.
Serum levels of IgM, IgG1, IgG2a, IgG2b, IgG3, and IgA were normal in 5-week-old CD81-deficient mice. In contrast, at 14 weeks of age, serum IgM and IgA levels in CD81-deficient mice were significantly increased 3 to 4 fold over those of wild-type littermates. Enhanced responses to natural type II TI antigens such as those derived from bacteria that colonize the gut may account for the age-dependent elevation of serum IgM and IgA in CD81-deficient mice.
Our findings that CD81 is not necessary for T cell development but is required for the expression of normal levels of CD19 are in agreement with a recent report by Maecker et al. (54). However, these authors found normal B-1 cells and decreased antibody response to the TD antigen ovalbumin in CD81-deficient mice. Our finding of enhanced antibody responses to type II TI antigens in CD81-deficient mice, taken together with the impaired TD antibody responses in CD19-deficient mice suggest, that CD19 may exert dual effects on BCR signaling: an enhancing effect mediated by its intracellular domain and an inhibitory mediated by CD81. It is tempting to suggest that the inhibitory signal may be dependent CD81-mediated calcium transport.
Acknowledgments
We thank Paul Swinton for technical assistance, Dr. Jonathan Spergel for the ELISA, and Drs. Fred Alt, Cox Terhorst, Hans Oettgen, and Francisco A. Bonilla for critical reading of the manuscript. We also thank Drs. Shihichi Sato and Thomas F. Tedder for their generous gift of anti-CD19 mAb MB19-1. This work was supported by National Institutes of Health Grant HD 17461 and by grants from Baxter Heathcare, Alpha Therapeutics, and Olsten Corporation.
ABBREVIATIONS
- [Ca2+]i
intracellular free calcium
- TM4SF
transmembrane-4 superfamily
- BCR
B cell antigen receptor
- TD
thymus dependent
- TI
T independent
- ES cells
embryonic stem cells
- TK
thymidine kinase
- TNP
2,4,6,-trinitrophenyl
- KLH
keyhole limpet hemocyanin
- FITC
fluorescein isothiocyanate
- LPS
lipopolysaccaride
- PE
phycoerythrin
References
- 1.Oren R, Takahashi S, Doss C, Levy R, Levy S. Mol Cell Biol. 1990;10:4007–4015. doi: 10.1128/mcb.10.8.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagira M, Imai T, Ishikawa I, Uwabe K I, Yoshie O. Cell Immunol. 1994;157:144–157. doi: 10.1006/cimm.1994.1212. [DOI] [PubMed] [Google Scholar]
- 3.Wright M D, Tomlison M G. Immunol Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 4.Tedder T F, Inaoki M, Sato S. Immunity. 1997;6:107–118. doi: 10.1016/s1074-7613(00)80418-5. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto A K, Martin D R, Carter R H, Klickstein L B, Ahearn J M, Fearon D T. J Exp Med. 1993;178:1407–1417. doi: 10.1084/jem.178.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradbury L E, Kansas G S, Levy S, Evans R L, Tedder T F. J Immunol. 1992;149:2841–2850. [PubMed] [Google Scholar]
- 7.Bradbury L E, Goldmacher V S, Tedder T F. J Immunol. 1993;151:2915–2927. [PubMed] [Google Scholar]
- 8.Carter R H, Fearon D T. Science. 1992;256:105–107. [PubMed] [Google Scholar]
- 9.Carroll M C, Fischer M B. Curr Opin Immunol. 1997;9:64–69. doi: 10.1016/s0952-7915(97)80160-4. [DOI] [PubMed] [Google Scholar]
- 10.Dempsey, P. W., Allison, M. E. D., Akkaraju, S., Goodnow, C. C. & Fearon, D. T. (271) Science 271, 348–350. [DOI] [PubMed]
- 11.Ahearn J M, Fischer M B, Croix D, Goerg S, Ma M, Xia J, Zhou X, Howard R G, Rothstein T L, Carrol M C. Immunity. 1996;4:251–262. doi: 10.1016/s1074-7613(00)80433-1. [DOI] [PubMed] [Google Scholar]
- 12.Croix D A, Ahearn J M, Rosengard A M, Han S, Kelsoe G, Ma M, Carrol M C. J Exp Med. 1996;183:1857–1864. doi: 10.1084/jem.183.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engel P, Zhou L J, Ord D C, Sato S, Koller B, Tedder T F. Immunity. 1995;3:39–50. doi: 10.1016/1074-7613(95)90157-4. [DOI] [PubMed] [Google Scholar]
- 14.Molina H, Holers V M, Li B, Fang Y F, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr R W, Chaplin D D. Proc Natl Acad Sci USA. 1996;93:3357–3361. doi: 10.1073/pnas.93.8.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickert R C, Rajewsky K, Roes J. Nature (London) 1995;376:352–355. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- 16.Imai T, Yoshie O. J Immunol. 1993;151:6470–6481. [PubMed] [Google Scholar]
- 17.Imai T, Kakizaki M, Nishimura M, Yoshie O. J Immunol. 1995;155:1229–1239. [PubMed] [Google Scholar]
- 18.Boismenu R, Rhein M, Fischer W H, Havran W L. Science. 1996;271:198–200. doi: 10.1126/science.271.5246.198. [DOI] [PubMed] [Google Scholar]
- 19.Schick M R, Levy S. J Immunol. 1993;151:4090–4097. [PubMed] [Google Scholar]
- 20.Rubenstein E, Le Naour F, Lagaudrere-Gesbert C, Billard M, Conjeaud H, Boucheix C. Eur J Immunol. 1996;26:2657–2665. doi: 10.1002/eji.1830261117. [DOI] [PubMed] [Google Scholar]
- 21.Angelisova P, Hilgert I, Horejsi V. Immunogenetics. 1994;39:249–256. doi: 10.1007/BF00188787. [DOI] [PubMed] [Google Scholar]
- 22.Radford K J, Thorne R F, Hersey P. Biochem Biophys Res Commun. 1996;222:13–18. doi: 10.1006/bbrc.1996.0690. [DOI] [PubMed] [Google Scholar]
- 23.Mannion B A, Berditchevski F, Kraeft S K, Chen L B, Hemler M E. J Immunol. 1996;157:2039–2047. [PubMed] [Google Scholar]
- 24.Berditchevski F, Zutter M M, Hemler M E. Mol Biol Cell. 1996;7:193–201. doi: 10.1091/mbc.7.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeyama S M, Koyama M, Yamaoko M, Sasada R, Miake M. J Exp Med. 1993;177:1231–1235. doi: 10.1084/jem.177.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garofalo A, Chifivi G S, Foglieni C, Pigott R, Mortenini R, Martin-Padura I, Anichini A, Gearing A J, Sanchez-Madrid F, Dejana E, Giavazzi R. J Exp Med. 1995;182:1191–1198. [PubMed] [Google Scholar]
- 27.Qian F, Vaux D L, Weissman I L. Cell. 1995;77:335–342. doi: 10.1016/0092-8674(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 28.Radford K J, Mallesh J, Hersey P. Int J Cancer. 1995;62:631–638. doi: 10.1002/ijc.2910620523. [DOI] [PubMed] [Google Scholar]
- 29.Hollander G A, Castigli E, Kulbacki R, Su M, Burakoff S J, Gutierrez-Ramos J C, Geha R S. Proc Natl Acad Sci USA. 1996;93:4994–4999. doi: 10.1073/pnas.93.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greimers R, Trebak M, Moutschen M, Boniver J. Cytometry. 1996;23:205–217. doi: 10.1002/(SICI)1097-0320(19960301)23:3<205::AID-CYTO4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Hayakawa K, Hardy R R, Herzenberg L A, Herzenberg L A. J Exp Med. 1985;161:1554–1559. doi: 10.1084/jem.161.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy R R, Li Y S, Hayakama K. Semin Immunol. 1996;8:37–44. doi: 10.1006/smim.1996.0006. [DOI] [PubMed] [Google Scholar]
- 33.Herzenberg L A, Haughton G, Rajewsky K. CD 5 B Cells in Development and Disease. New York: N.Y. Acad. Sci.; 1992. [Google Scholar]
- 34.Tarakhovsky A, Turner M, Schaal S, Mee P J, Duddy L P, Rajewsky K, Tybulewicz V L. Nature (London) 1995;374:467–470. doi: 10.1038/374467a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang R, Alt F W, Davidson L, Orkin S H, Swat W. Nature (London) 1995;374:470–473. doi: 10.1038/374470a0. [DOI] [PubMed] [Google Scholar]
- 36.Khan W N, Alt F W, Gerstein R M, Malynn B A, Larsson I, Rathbun G, Davidson L, Muller S, Kantor A B, Herzenberg L A, Rosen F S, Sideras P. Immunity. 1995;3:283–299. doi: 10.1016/1074-7613(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 37.Leitges M, Schmedt C, Gunamard R, Davoust J, Schaal S, Stabel S, Tarakhovsky A. Science. 1996;273:788–791. doi: 10.1126/science.273.5276.788. [DOI] [PubMed] [Google Scholar]
- 38.Kozlowski M, Mlinaric-Rscan I, Feng G S, Shen R, Pawson T, Siminovitch K A. J Exp Med. 1993;178:2157–2163. doi: 10.1084/jem.178.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulte R J, Campbell M A, Fischer W H, Sefton B M. Science. 1992;258:1001–1004. doi: 10.1126/science.1279802. [DOI] [PubMed] [Google Scholar]
- 40.O’Keef T L, Williams G T, Davies S L, Neuberger M S. Science. 1996;274:798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 41.Sato S, Miller A S, Inaoki M, Bock C B, Jansen P J, Tang M L K, Tedder T F. Immunity. 1996;5:551–562. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 42.Sato S, Ono N, Steeber D A, Pisetsky D S, Tedder T F. J Immunol. 1996;157:4371–4378. [PubMed] [Google Scholar]
- 43.Krop I, de Fougerolles A R, Hardy R R, Alison M, Schlissel M S, Fearon D T. Eur J Immunol. 1996;26:238–242. doi: 10.1002/eji.1830260137. [DOI] [PubMed] [Google Scholar]
- 44.Pezzutto A, Dorken B, Rabinovich P S, Ledbetter J A, Moldenhauer G, Clark E A. J Immunol. 1987;138:2793–2799. [PubMed] [Google Scholar]
- 45.Carter R H, Tuveson D A, Park D J, Rhee S G, Fearon D T. J Immunol. 1991;147:366–371. [PubMed] [Google Scholar]
- 46.Gil M L, Vita N, Lebel-Binay S, Miloux B, Chalon P, Kaghad M, Marchiol-Foupnigault C, Conjeaud H, Caput D, Ferrara P, Fradelizi D, Quillet-Mary A. J Immunol. 1992;148:2826–2833. [PubMed] [Google Scholar]
- 47.Tedder T F, Streuli M, Schlossman S F, Saito H. Proc Natl Acad Sci USA. 1988;85:208–212. doi: 10.1073/pnas.85.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bubien J K, Zhou L J, Bell P D, Frizzell R A, Tedder T F. J Cell Biol. 1993;121:1121–1132. doi: 10.1083/jcb.121.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unwin, N. (1993) Cell 10, Suppl., 31–41.
- 50.Rigley K P, Callard R E. Eur J Immunol. 1991;21:535–540. doi: 10.1002/eji.1830210302. [DOI] [PubMed] [Google Scholar]
- 51.Ledbetter J A, Rabinovich P S, June C H, Song C H, Clark E A, Uckun F M. Proc Natl Acad Sci USA. 1988;85:1897–1901. doi: 10.1073/pnas.85.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato S, Steeber D A, Tedder T F. Proc Natl Acad Sci USA. 1995;92:11558–11562. doi: 10.1073/pnas.92.25.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato S, Steeber D A, Jansen P J, Tedder T F. J Immunol. 1997;158:4662–4669. [PubMed] [Google Scholar]
- 54.Maecker H T, Levy S. J Exp Med. 1997;185:1505–1510. doi: 10.1084/jem.185.8.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]