Abstract
Although cellular proteins degraded by proteasomes are the source of most antigenic peptides presented on major histocompatibility complex class I molecules, it is unknown whether the eight- to nine-residue peptides that fit in the binding groove of class I molecules are directly produced by proteasomes alone in vivo. If the eight-residue peptide SIINFEKL from chicken ovalbumin is extended by one or several residues at its C terminus and microinjected into cells or expressed from a minigene, it is processed and presented on major histocompatibility complex class I. However, processing and presentation are inhibited by proteasome inhibitors, such as lactacystin. In contrast, when SIINFEKL is extended by 2 to 25 residues at its N terminus, its presentation is not blocked by proteasome inhibitors. N-terminal processing also can occur when the extended peptide is cotranslationally inserted into the endoplasmic reticulum. Thus, two different proteolytic steps in the generation of an chicken ovalbumin-presented peptide can be distinguished. Cleavage by the proteasome defines the proper C terminus, whereas distinct peptidase(s) in the cytosol or endoplasmic reticulum may generate the appropriate N terminus from extended peptides.
The continual presentation of fragments of intracellular proteins on major histocompatibility complex (MHC) class I molecules is a process that allows T lymphocytes to identify and selectively eliminate those cells that are synthesizing foreign or abnormal proteins (1). In this process, oligopeptides are generated during the degradation of proteins, and transported from the cytosol into the endoplasmic reticulum (ER) where they bind to newly synthesized class I molecules. These complexes with bound antigenic peptides then are transported to the plasma membrane, where they are displayed to T cells (1). The peptides bound to MHC class I molecules are remarkably uniform in size, typically eight or nine residues. Only peptides of this length are tightly bound, because the class I peptide-binding groove has closed ends, and the interaction of peptide N and C termini with pockets in the MHC molecule contributes significantly to the overall affinity of binding (1–4). The present studies were undertaken to clarify the mechanisms that are responsible for generating peptides of this precise size for presentation.
It is now well established that generation of most MHC class I-presented peptides requires the 20S proteasome, a 700-kDa particle with multiple peptidase activities (5–9). The 20S proteasome functions as the proteolytic core of a 26S (2,000 kDa) complex that degrades ubiquitinylated proteins (5, 6, 8, 10, 11). It remains unclear, however, whether proteasomes alone catalyze the production of eight- to nine-residue MHC-bound peptides. Information on the sizes of peptides produced by mammalian 20S or 26S proteasomes during protein degradation in vivo is not available. Although the simpler Archaebacterial 20S proteasome, which contains only one type of active site, generates peptides averaging 7–8 residues, the range of peptide products varies widely in size, from five to 20 residues (T.A. and A.G., unpublished observations). Purified 20S mammalian proteasomes incubated for extended periods with chicken ovalbumin (OVA) or β-galactosidase, under highly nonphysiological conditions, can generate some of the antigenic peptides presented in vivo, but also can generate many other products, including longer peptides containing internal epitope sequences (12–14). It remains unclear whether these findings are relevant to the degradative process functioning in vivo, when proteolysis involves ubiquitin and 26S complexes, and where other peptidases can rapidly degrade peptide products released by proteasomes (6, 8, 10, 11).
Recently, specific inhibitors of proteasomes, such as peptide aldehydes or lactacystin, have been described that can enter cells and inhibit proteasome function (7, 9, 15). These agents block the degradation of the majority of cellular proteins and prevent the generation of most class I presented-peptides (7, 9). The inhibitors therefore were used in the present studies to determine whether proteasomes in vivo are likely to generate the N and C termini characteristic of a MHC-bound epitope, the OVA-derived peptide SIINFEKL. In addition, we tested whether longer peptides that may be generated by the proteasome can be processed into the presented peptide epitope by proteasomal or other cellular proteolytic systems.
MATERIALS AND METHODS
Plasmid Constructs.
All constructs expressed genes under control of the T7 RNA polymerase promoter in pBluescript SK (Promega). The plasmid p.OVA was constructed by cloning OVA cDNA into pBluescript SK, using HindIII and XbaI (all enzymes from Life Technologies or New England Biolabs). Other constructs (except p.25+SIINFEKL) were prepared by phosphorylation and annealing of complementary oligonucleotide primers (synthesized at Genosys, The Woodlands, TX or the Dana–Farber Cancer Institute Molecular Biology Core Facility, Boston, MA) with restriction site overhangs (HindIII at the 5′ end and EcoRI at the 3′ end), then ligation into pBluescript SK (see Table 1 for translated products of expressed genes). The coding primer consisted of a Kozak consensus sequence and start codon followed by the relevant DNA sequence, two stop codons, and an NcoI site for selection. The p.25.SIINFEKL coding primer had a HindIII 5′ overhang, a StuI 3′ overhang, and a BglII site for selection 5′ of the Kozak consensus sequence, and was ligated into the p.15.SIINFEKL plasmid. Maxipreps generally were performed with QIAfilter MAXI kits (Qiagen, Chatsworth, CA), and purity was assessed by spectrophotometry and on agarose gels. cDNAs were sequenced at the Dana–Farber Cancer Institute Molecular Biology Core Facility. Amino acid sequences for the translated products of plasmid constructs are noted in Table 1.
Table 1.
Translated products of expressed genes and synthetic peptide sequences
| Name | Amino acid sequence* |
|---|---|
| Plasmid | |
| p.SIINFEKL | SIINFEKL |
| p.5+SIINFEKL+5 | LEQLESIINFEKLTEWTS |
| p.SIINFEKL+15 | SIINFEKLTEWTSSNVMEERKIK |
| p.SIINFEKL+5 | SIINFEKLTEWTS |
| p.SIINFEKL+4 | SIINFEKLTEWT |
| p.SIINFEKL+3 | SIINFEKLTEW |
| p.SIINFEKL+2 | SIINFEKLTE |
| p.SIINFEKL+1 | SIINFEKLT |
| p.25+SIINFEKL | LPFASGTMSMLVLLPDEVSGLEQLESIINFEKL |
| p.15+SIINFEKL | LVLLPDEVSGLEQLESIINFEKL |
| p.5+SIINFEKL | LEQLESIINFEKL |
| p.2+SIINFEKL | LESIINFEKL |
| p.ss.SIINFEKL | †SIINFEKL |
| p.ss.5+SIINFEKL | †LEQLESIINFEKL |
| p.ss.SIINFEKL+5 | †SIINFEKLTEWTS |
| Peptide | |
| SIINFEKL | SIINFEKL |
| 8+SIINFEKL+8 | VSGLEQLESIINFEKLTEWTSSNV |
| SIINFEKL+5 | SIINFEKLTEWTS |
| 3+SIINFEKL | QLESIINFEKL |
SIINFEKL is encoded by codons 257–264 of OVA cDNA.
Sequences are of translated minigene products.
Sequence is preceded by the e3/19 K signal sequence.
Virus Stock and Synthetic Peptides.
vTF7–3 recombinant vaccinia virus was obtained from American Type Culture Collection (VR-2153) and propagated in human 143B TK− cells (ATCC CRL 8303). Peptides SIINFEKL, 8+SIINFEKL+8, SIINFEKL+5, and 3+SIINFEKL were synthesized and HPLC-purified at the molecular biology core facility of the Dana–Farber Cancer Institute. Synthetic peptides used as standards in HPLC fractionation assays were manufactured by Macromolecular Resources (Colorado State University, Fort Collins, CO).
Cell Lines and Antigen Presentation Assays.
Antigen-presenting cells (APCs) were: E36.12.4, a hamster lung carcinoma cell line stably transfected with H-2 Kb and ICAM-1 (16); DAP34.8, a similarly transfected murine L cell line; LB27.4, murine B lymphoblastoid cells (17); or BM K07 transporter associated with antigen presentation (TAP) (−/−) cells, immortalized from bone marrow of TAP (−/−) mice by transduction of myc and raf (kindly provided by M. Kovacsowicz-Bankowski, Dana-Farber Cancer Institute). Cells were passaged in RPMI 1640 medium or DMEM with 10% fetal calf serum, in the presence of gentamycin for stably transfected cells, or in Optimem with 1% normal mouse serum (for cytosolic loading assays) (18). Assays were performed essentially as described (7, 9, 18). APCs were infected with vTF7–3 and then transfected by lipofection with plasmid-encoding antigen, or were cytosolically loaded with peptide by electroporation (7, 9, 18). After 140 min at 37°C, APCs were fixed in 1% paraformaldehyde, and the presence of peptide-MHC complexes was assayed by measuring the production of interleukin 2 from RF33.70 T-T hybridomas (specific for SIINFEKL-Kb) (18).
Cell Extracts and HPLC Fractionation.
Total acid-soluble peptide pools were prepared according to the method of Malarkannan et al. (19). E36.12.4 cells (30 × 106) were extracted with trifluoroacetic acid 24 hr after transfection with 40 μg antigen-expressing plasmid. The low molecular weight material was fractionated using a Hewlett-Packard HPLC system. Reverse-phase C18 columns were run in 0.05% trifluoroacetic acid (TFA) in water (solvent A) and 0.06% TFA in 80% acetonitrile (solvent B), with a gradient for separations of 25% to 55% over 30 min, with fractions collected every minute. Dilutions (1:20) of fractions were added to 105 fixed E36.12.4 cells and assayed for the ability to stimulate RF33.70 cells.
RESULTS
Presentation of SIINFEKL from Minigenes and Synthetic Peptides.
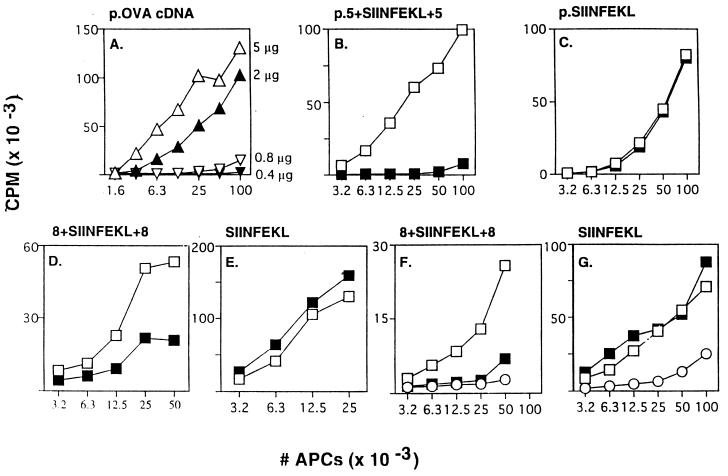
The antigenic epitope derived from OVA is the peptide SIINFEKL (amino acids 257–264), which binds to murine class I Kb. To analyze the proteolytic steps involved in generating this epitope we first explored whether cells could use a smaller region of OVA for antigen presentation, which then could be truncated further to clarify the subsequent cleavage steps in vivo. A minigene containing SIINFEKL with five additional residues of the OVA sequence extending from both its N and C termini (p.5+SIINFEKL+5) was inserted in a plasmid under the control of a T7 promoter and transfected into APCs expressing T7 polymerase from a recombinant vaccinia virus (vTF7–3). Presentation of SIINFEKL on Kb was determined using RF33.70 T-T hybridomas (19). With this experimental system, we have shown that the amount of antigen presented increases with the amount of plasmid transfected (Fig. 1A) and the length of time before fixation (data not shown).
Figure 1.
MHC class I antigen presentation from OVA protein and SIINFEKL-containing oligopeptides. (A) E36.12.4 APCs were infected with vTF7–3 and then transfected with the indicated concentrations of plasmid encoding intact OVA cDNA. Assays for presentation of SIINFEKL on Kb were performed as described (7). (B) Similar to A, except that cells were treated with (▪) or without (□) lactacystin (2 μM) for 30′ before transfection and during the subsequent incubation, and 5 μg of a plasmid encoding p.5+SIINFEKL+5 was transfected. (C) Same as B except that the plasmid encoded p.SIINFEKL (0.7 μg transfected). (D) Similar to B except that 8+SIINFEKL+8 synthetic peptide (300 μg/ml) was introduced into the cytosol of LB27.4 cells (17) (APCs) by electroporation (7) (instead of vaccinia infection and plasmid transfection) and lactacystin was used at 40 μM. (E) Same as D except that SIINFEKL (0.5 μg/ml) was used instead of 8+SIINFEKL+8 peptide. (F) Same as D except that LLnL (40 μM) was used instead of lactacystin. (G) Same as E except LLnL (40 μM) was used instead of lactacystin. Some APCs were fixed immediately after electroporation of antigen (○) to rule out peptide binding directly to cell surface MHC molecules (7). All results in Figs. 1, 2, 3, 4 are representative of data obtained in repeated experiments. OVA constructs expressed from plasmids are indicated with the prefix p.
The class I presentation of antigenic peptides from OVA is dependent on degradation by the proteasome, as shown by sensitivity to various proteasome-specific inhibitors, including peptide aldehydes and lactacystin (7, 9). We therefore examined whether the presentation of the p.5+SIINFEKL+5 construct (see Table 1 for expressed sequences) was affected by the inhibitor lactacystin (9, 15). Lactacystin did inhibit the presentation of the extended construct (Fig. 1B), which requires proteolysis for the generation of SIINFEKL. In contrast, lactacystin did not inhibit presentation from a construct encoding the SIINFEKL epitope alone (Fig. 1C). These results indicate that lactacystin selectively interfered with the proteolytic generation of SIINFEKL from the larger construct and did not interfere with other steps in the antigen-presentation pathway. Similarly, lactacystin did not block antigen presentation when synthetic SIINFEKL peptide was introduced into APCs by electroporation (Fig. 1E), but did block presentation of a SIINFEKL peptide extended by eight residues at both termini (8+SIINFEKL+8) (Fig. 1D). Furthermore, N-acetyl-l-leucinyl-l-leucinal-l-norleucinal (LLnL) (7), a peptide aldehyde inhibitor of the proteasome, also inhibited presentation of 8+SIINFEKL+8 synthetic peptide (Fig. 1F) and 5+SIINFEKL+5 expressed from a minigene (data not shown), without affecting the presentation of SIINFEKL itself (Fig. 1G) (see Table 1 for synthetic peptide sequences).
In general, inhibition of SIINFEKL presentation from extended constructs was strong but not as complete as inhibition from expressed intact OVA, probably because many cleavages are necessary to generate the epitopes from whole protein substrates. The effects of inhibitors on whole proteins therefore would be more distinct than effects on extended peptide constructs, which require one or a few cleavages to generate SIINFEKL. Alternatively, there may be some distinct, but minor, pathway involved in peptide generation. Previously, others have used similar extended peptide constructs in cell-free degradation systems to study peptide generation by proteasomes, without establishing whether presentation was dependent on proteasome activity in vivo (12–14). Our results indicate that the generation in vivo of SIINFEKL from the extended constructs is largely dependent on the proteasome. We therefore could truncate the extended construct further to define the actual sites cleaved by proteasomes.
Analysis of Proteasomal Involvement in Generating SIINFEKL N and C Termini.
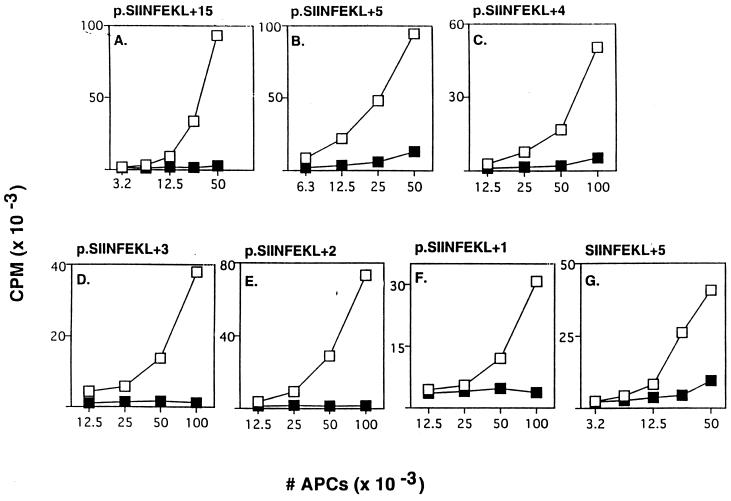
To examine whether the proteasome was involved in trimming the C-terminal flanking residues of SIINFEKL, minigenes consisting of SIINFEKL extended by 15 (Fig. 2A) or 5 (Fig. 2B) residues at its C terminus (p.SIINFEKL+15 and p.SIINFEKL+5) were expressed in APCs. The presentation of SIINFEKL from these constructs was inhibited by lactacystin. Identical results were obtained when a synthetic SIINFEKL+5 peptide was electroporated into APCs (Fig. 2G). These results indicate that the proteasome is responsible for cleavage(s) somewhere in the C-terminal extension region.
Figure 2.
Proteasome inhibitors block presentation of SIINFEKL with C-terminal extensions. (A-F) Similar to Fig. 1B except the indicated plasmids (5 μg) encoding SIINFEKL with 15, 5, 4, 3, 2, or 1 C-terminal flanking residues were used, with 0 μM (□) or 2 μM (▪) lactacystin. (G) Similar to Fig. 1D except SIINFEKL+5 peptide (25 μg/ml) was used. Data are from independent experiments.
To map precisely where the proteasome might be cutting the C-terminal extension, we expressed minigenes encoding SIINFEKL with an extension of 4, 3, 2, or 1 residues at the C terminus in APCs. The presentation of all of these constructs was blocked similarly by lactacystin (Fig. 2 C-F) and LLnL (data not shown). Because proteasome activity was required for presentation of the p.SIINFEKL+1 construct (Fig. 2F), in vivo proteasomes must be responsible for generating the precise C-terminal end of the SIINFEKL epitope.
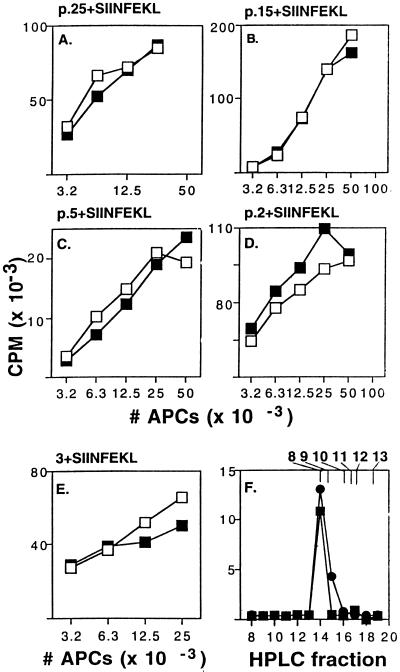
We next examined whether the proteasome also was required for trimming the N-terminal flanking residues of the SIINFEKL peptide. SIINFEKL minigene constructs with 25-, 15-, 5-, or 2-residue N-terminal extensions were expressed in APCs (Fig. 3 A-D). Surprisingly, neither lactacystin (Fig. 3) nor LLnL reduced the presentation of any of the N-terminal extended constructs, at concentrations that maximally inhibited degradation of cellular proteins and presentation from intact OVA or C-terminal-extended constructs. Similarly, when a synthetic SIINFEKL peptide extended by three residues on its N terminus (3+SIINFEKL) was electroporated into APCs, lactacystin did not inhibit presentation (Fig. 3E). Interestingly, the presentation of these constructs typically required the introduction of 5-fold less plasmid into cells than was required for presentation from transfects with OVA cDNA or SIINFEKL with C-terminal flanking residues. These findings suggest that processing of the N-terminal extension is more efficient than processing of the C-terminal flank.
Figure 3.
Proteasome inhibitors do not affect presentation of SIINFEKL with N-terminal extensions. (A-D) Similar to Fig. 2 except the indicated plasmids (1.5 μg) with 2, 5, 15, or 25 N-terminal flanking residues were used, with 0 μM (□) or 2 μM (▪) lactacystin. Similar results were obtained using 20 μM lactacystin (not shown). (E) Similar to Fig. 2G except 3+SIINFEKL peptide (5 μg/ml) was used. Data are from independent experiments. (F) Extracts from cells transfected with plasmid p.5+SIINFEKL (▪) or p.SIINFEKL (•) were fractionated, and RF33.70-stimulatory capacity of fractions was assayed. The elution profile of synthetic peptides corresponding to SIINFEKL with 0, 1, 2, 3, 4, and 5 N-terminal flanking residues are indicated at the top.
To confirm that the N-terminal extensions on SIINFEKL were actually being trimmed in vivo, before presentation on MHC molecules, we analyzed the Kb-presented peptides that were generated from these constructs. p.5+SIINFEKL was expressed in APCs, and then peptides were extracted in trifluoroacetic acid and fractionated by reverse-phase HPLC (19). The only fraction with stimulatory activity for RF33.70 T hybridomas was fraction 14, the same fraction in which the synthetic SIINFEKL 8-mer peptide elutes (Fig. 3F, squares). Cell extracts from cells transfected with p.2+SIINFEKL and p.SIINFEKL alone (Fig. 3F, circles) also had activity only in fraction 14. This HPLC system could resolve SIINFEKL itself from SIINFEKL extended by 1–5 residues on its N terminus (Fig. 3F). These results indicate that the N-terminal extensions are trimmed in APCs to generate the appropriate octamer peptide, and that this process does not require the proteasome.
Analysis of SIINFEKL Generation in Interferon γ (IFN-γ)-Treated Cells.
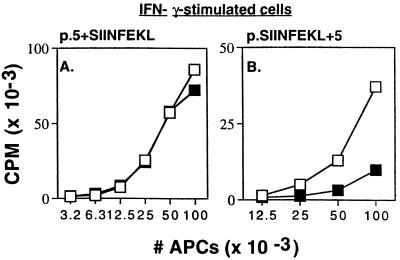
Treatment of cells with IFN-γ stimulates cellular expression of the proteasome catalytic β-subunits LMP2, LMP7, and MECL-1 (20–25), as well as PA28, a complex that enhances the 20S proteasome peptidase activities (26–28). These IFN-γ-induced proteins have been shown to enhance antigen presentation of certain model antigens (23, 24, 29) and to promote the generation of eight-residue peptides from longer peptides in cell-free systems (12). Therefore, it is possible that proteasomes in IFN-γ-stimulated cells also become primarily responsible for generating the correct peptide N termini. We therefore examined the presentation of N- and C-terminal extended SIINFEKL constructs in IFN-γ-stimulated cells. We found that lactacystin still inhibited presentation of the p.SIINFEKL+5 construct but did not affect presentation of the p.5+SIINFEKL construct (Fig. 4 A and B), as was found in control cells. IFN-γ-induced subunits favor production of C-terminal residues appropriate for binding to class I molecules (see below). However, even in the presence of IFN-γ other enzymes seem to generate the characteristic N-terminal residues.
Figure 4.
Class I presentation from extended SIINFEKL constructs in IFN-γ-stimulated cells. Antigen presentation assays using (A) p.5+SIINFEKL plasmid or (B) p.SIINFEKL+5 were performed as described in Fig. 2, except that the APCs were IFN-γ-stimulated DAP34.8 cells with (▪) or without (□) 3 μM lactacystin. Cells were incubated for 3 days at 37°C in the presence or absence of murine IFN-γ (from supernatants of IFN-γ-transduced B16 cells, kindly provided by Glenn Dranoff, Dana–Farber Cancer Institute, containing approximately 10 units/ml IFN-γ). Lactacystin had the same pattern of effects in nonstimulated DAP34.8.
N-Terminal Trimming in the ER.
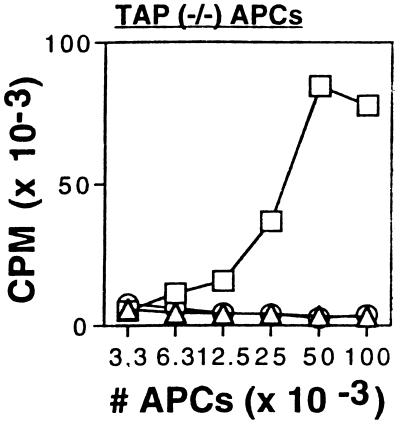
The N-terminal trimming of these extended peptides in principle might occur in the cytoplasm and/or the ER. To evaluate the ability of the ER to trim the N-terminal regions, we fused the signal sequence from adenovirus E3/19K to our constructs (30). The expressed peptides are cotranslationally transported into the ER where the signal sequence then is removed. To exclude any possible contribution from proteolysis in the cytoplasm we expressed these constructs in cells that lacked functional TAP, which is required to deliver antigenic peptides generated in the cytosol into the ER. The ER-targeted SIINFEKL with a five-residue N-terminal extension (p.ss.5+SIINFEKL) was presented efficiently (Fig. 5, □), and presentation was not inhibited by lactacystin (not shown). In contrast, ER-targeted SIINFEKL with a five-residue C-terminal extension (p.ss.SIINFEKL+5) was not presented in TAP-deficient cells (Fig. 5, ▵), even at high concentrations, similarly to a SIINFEKL construct lacking a signal sequence (Fig. 5, ○).
Figure 5.
Class I presentation from extended SIINFEKL constructs targeted to the ER. Antigen presentation assays were performed as described in Fig. 1A using plasmid constructs encoding 5+SIINFEKL (5 μg) (○), 5+SIINFEKL with an adenovirus E3/19K ER targeting sequence at the N terminus (2 μg) (□) or ER-targeted SIINFEKL+5 (5 μg) (▵). APCs were TAP-deficient murine BM K07 cells.
DISCUSSION
These results demonstrate that two distinct proteolytic steps can function in the generation of a presented peptide. One step is performed by the proteasome. We show in vivo that this particle is directly responsible for the cleavage(s) that produces the exact C-terminal end of a presented peptide. It previously had been proposed that the proteasome was responsible for the C-terminal cleavage because the IFN-γ-inducible proteasome β-subunits (LMP2 and 7) promoted cleavages after hydrophobic and basic residues while reducing hydrolysis after acidic residues (21, 22). Therefore, more peptides should be generated that end with hydrophobic and basic residues, which are preferentially transported into the ER and bind much more efficiently to MHC class I molecules. Moreover, these changes in cleavage specificity did not correlate with the nature of the cleavages generating the N termini. Our results demonstrate that the precise C-terminal cleavage for SIINFEKL is produced in vivo by the proteasome, and presumably proteasomes containing LMP2 and seven subunits can perform this cleavage more efficiently.
Our findings also suggest that there is little or no carboxypeptidase activity in the cytoplasm, or at least insufficient activity for trimming of C-terminal residues in the absence of proteasome activity. Alternatively, the C-terminal residues of peptides may be somehow protected from these activities, perhaps by chaperones. Because it is likely that C-terminal extended peptides are transported into the ER by TAP but do not present SIINFEKL on class I, this implies there is probably also little carboxypeptidase activity in the ER. Earlier findings also had suggested that the ER had little or no ability for trimming the C terminus of peptide (31–33), supported by our finding that SIINFEKL with a C-terminal extension is not processed in the ER (Fig. 5). In one study, peptides with C-terminal extensions that were delivered into this compartment were not presented well unless a carboxypeptidase was coexpressed (32). Similarly, a polymorphism in the rat TAP molecule inhibited the transport of peptides terminating in Arg and thereby prevented presentation of such peptides (34); this indirectly argued that new C-terminal residues (some of which would contain Arg) were not generated in the ER. The potential for C-terminal trimming in the cytosol has not previously been examined. If carboxypeptidase activity is generally low in these intracellular compartments, then proteasomes are probably responsible for determining the exact C-terminal residues of most antigenic peptides. (However, this model will need to be formally examined and exceptions are likely to exist (35, 36.)
We demonstrate a second proteolytic step that trims the N-terminal end of a peptide. This step is resistant to lactacystin at concentrations that maximally inhibit proteasome-dependent degradation of long- and short-lived proteins in vivo (9) as well as the presentation of SIINFEKL from intact whole OVA and from SIINFEKL with C-terminal extensions. Because lactacystin inactivates all catalytically active subunits of proteasomes (9) it is likely that this second step is mediated by a distinct protease, most likely an aminopeptidase. This trimming step appears to be efficient because the presentation of SIINFEKL constructs with N-terminal extensions requires five times lower concentrations of plasmid than constructs with C-terminal overhangs. A similar difference in efficiency of presentation was noted when the analogous peptides were electroporated into cells. Although proteasomes may make the correct N- and C-terminal cleavages when hydrolyzing a full-length protein, they are likely to also make many longer oligopeptides (12–14). Therefore, the trimming step(s) defined here would allow these extended products to be efficiently presented.
Our results, together with earlier studies that directed extended peptides into the ER (30–33), demonstrate that N-terminal trimming can occur in this compartment, presumably through aminopeptidases, although signal peptidase also may participate in the removal of residues directly following the leader sequence. The TAP transporter has preference for certain C termini but can translocate peptides longer than eight or nine residues, which would require trimming for presentation. However, because peptides bind to class I molecules associated with TAP immediately after translocation into the ER there may be little opportunity for trimming to occur in this compartment. In general, there appears to be little trimming of peptides after binding to class I molecules (37). However, it is possible that N-terminal trimming might allow the presentation of longer peptides that are translocated into the ER and fail to bind stably to class I molecules.
The TAP transporter translocates peptides longer than 15 residues inefficiently in vitro (38, 39). Our finding that SIINFEKL constructs with a 25-residue N-terminal extension (32 residues in total) present SIINFEKL as efficiently as shorter constructs suggests that they are trimmed in the cytoplasm before transport. Aminopeptidases that are candidates for this activity exist in the cytoplasm. Interestingly, at least one of these, leucine aminopeptidase, is induced by IFN-γ, a potent stimulator of class I antigen presentation (40). However, further experiments will be needed to prove such a role and to identify other cytosolic peptidases that process N-terminal extensions for antigen presentation.
A critical role of protein degradation in cells is to recycle proteins into amino acids for use in new synthesis or for energy metabolism (8, 10, 11). The proteasome plays a major role in this process by degrading the bulk of proteins into oligopeptides. The proteolytic activities that convert these oligopeptides to amino acids are not known. An important implication of our data is that this activity is likely to be mediated by aminopeptidases and possibly endopeptidases, with little if any role for carboxypeptidases.
Acknowledgments
We thank Alexei Kisselev for performing HPLC fractionations on cell extracts, and ProScript, Inc. (Cambridge, MA) for their generous gift of lactacystin. This work was supported by grants from the National Institute for Allergy and Infectious Disease for K.L.R. and from the National Institute of General Medical Science and Hieron Frontiers Science Program for A.G.
ABBREVIATIONS
- MHC
major histocompatibility complex
- OVA
chicken ovalbumin
- ER
endoplasmic reticulum
- APC
antigen-presenting cell
- LLnL
N-acetyl-l-leucinyl-l-leucinal-l-norleucinal
- IFN-γ
interferon γ
- TAP
transporter associated with antigen presentation
References
- 1.York I A, Rock K L. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Nature (London) 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 3.Madden D R, Gorga J C, Strominger J L, Wiley D C. Nature (London) 1991;353:321–325. doi: 10.1038/353321a0. [DOI] [PubMed] [Google Scholar]
- 4.Bouvier M, Wiley D C. Science. 1994;265:398–402. doi: 10.1126/science.8023162. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg A L, Rock K L. Nature (London) 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 6.Rivett J. Biochem J. 1993;291:1–10. doi: 10.1042/bj2910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg K L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 8.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–818. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 9.Craiu A, Gaczynska M, Akopian T, Gramm C, Fenteany G, Goldberg A L, Rock K L. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 11.Rubin D M, Finley D. Curr Biol. 1995;5:854–858. doi: 10.1016/s0960-9822(95)00172-2. [DOI] [PubMed] [Google Scholar]
- 12.Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski U H, Kloetzel P M. J Exp Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick L R, Aldrich C, Jameson S C, Moomaw C R, Pramanik B C, Doyle C K, DeMartino G N, Bevan M J, Forman J M, Slaughter C A. J Immunol. 1994;152:3884–3894. [PMC free article] [PubMed] [Google Scholar]
- 14.Niedermann G, Butz S, Ihlenfeldt H G, Grimm R, Lucchiari M, Hoschutzky H, Jung G, Maier B, Eichmann K. Immunity. 1995;2:289–299. doi: 10.1016/1074-7613(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 15.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 16.Michalek M T, Grant E P, Gramm C, Goldberg A L, Rock K L. Nature (London) 1993;363:552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 17.Kappler J, White J, Wegmann D, Mustain F, Marrack P. Proc Natl Acad Sci USA. 1982;79:3604–3607. doi: 10.1073/pnas.79.11.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rock K L, Rothstein L, Gamble S. J Immunol. 1990;145:804–811. [PubMed] [Google Scholar]
- 19.Malarkannan S, Goth S, Buchholz D R, Shastri N. J Immunol. 1995;154:585–598. [PubMed] [Google Scholar]
- 20.Monaco J J, McDevitt H O. Hum Immunol. 1986;15:416–426. doi: 10.1016/0198-8859(86)90019-4. [DOI] [PubMed] [Google Scholar]
- 21.Driscoll J, Brown M G, Finley D, Monaco J J. Nature (London) 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 22.Gaczynska M, Rock K L, Goldberg A L. Nature (London) 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 23.Fehling H J, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, von Boehmer H. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 24.Van Kaer L, Ashton-Rickardt P G, Eichelberger M, Gaczynska M, Nagashima K, Rock K L, Goldberg A L, Doherty P C, Tonegawa S. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg A L, Gaczynska M, Grant E P, Michalek M, Rock K L. Cold Spring Harbor Symp Quant Biol. 1995;60:479–490. doi: 10.1101/sqb.1995.060.01.052. [DOI] [PubMed] [Google Scholar]
- 26.Ma C P, Slaughter C A, DeMartino G N. J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 27.Groettrup M, Ruppert T, Kuehn L, Seeger M, Standera S, Koszinowski U, Kloetzel P M. J Biol Chem. 1995;270:23808–23815. doi: 10.1074/jbc.270.40.23808. [DOI] [PubMed] [Google Scholar]
- 28.Dick T P, Ruppert T, Groettrup M, Kloetzel P M, Kuehn L, Koszinowski U H, Stevanovic S, Schild H, Rammensee H G. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 29.Groettrup M, Soza A, Eggers M, Kuehn L, Dick T P, Schild H, Rammensee H G, Koszinowski U H, Kloetzel P M. Nature (London) 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- 30.Anderson K, Cresswell P, Gammon M, Hermes J, Williamson A, Zweerink H. J Exp Med. 1991;174:489–492. doi: 10.1084/jem.174.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonifacino J S, Lippincott-Schwartz J. Curr Opin Cell Biol. 1991;3:592–600. doi: 10.1016/0955-0674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- 32.Eisenlohr L C, Bacik I, Bennink J R, Bernstein K, Yewdell J W. Cell. 1992;71:963–972. doi: 10.1016/0092-8674(92)90392-p. [DOI] [PubMed] [Google Scholar]
- 33.Snyder H L, Yewdell J W, Bennink J R. J Exp Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powis S J, Young L L, Joly E, Barker P J, Richardson L, Brandt R P, Melief C J, Howard J C, Butcher G W. Immunity. 1996;4:159–165. doi: 10.1016/s1074-7613(00)80680-9. [DOI] [PubMed] [Google Scholar]
- 35.Yang B, Hahn Y S, Hahn C S, Braciale T J. J Exp Med. 1996;183:1545–1552. doi: 10.1084/jem.183.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yellen-Shaw A, Eisenlohr L C. J Immunol. 1997;158:1727–1733. [PubMed] [Google Scholar]
- 37.Ojcius D M, Langlade-Demoyen P, Gachelin G, Kourilsky P. J Immunol. 1994;152:2798–2810. [PubMed] [Google Scholar]
- 38.Neefjes J J, Momburg F, Hämmerling G J. Science. 1993;261:769–771. doi: 10.1126/science.8342042. [DOI] [PubMed] [Google Scholar]
- 39.Androlewicz M J, Cresswell P. Immunity. 1994;1:7–14. doi: 10.1016/1074-7613(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 40.Harris C A, Hunte B, Krauss M R, Taylor A, Epstein L B. J Biol Chem. 1992;267:6865–6869. [PubMed] [Google Scholar]