Abstract
Keratinocyte growth factor (KGF) is a member of the fibroblast growth factor family. Portions of the gene encoding KGF were amplified during primate evolution and are present in multiple nonprocessed copies in the human genome. Nucleotide analysis of a representative sampling of these KGF-like sequences indicated that they were at least 95% identical to corresponding regions of the KGF gene. To localize these sequences to specific chromosomal sites in human and higher primates, we used fluorescence in situ hybridization. In human, using a cosmid probe encoding KGF exon 1, we assigned the location of the KGF gene to chromosome 15q15–21.1. In addition, copies of KGF-like sequences hybridizing only with a cosmid probe encoding exons 2 and 3 were localized to dispersed sites on chromosome 2q21, 9p11, 9q12–13, 18p11, 18q11, 21q11, and 21q21.1. The distribution of KGF-like sequences suggests a role for alphoid DNA in their amplification and dispersion. In chimpanzee, KGF-like sequences were observed at five chromosomal sites, which were each homologous to sites in human, while in gorilla, a subset of four of these homologous sites was identified; in orangutan two sites were identified, while gibbon exhibited only a single site. The chromosomal localization of KGF sequences in human and great ape genomes indicates that amplification and dispersion occurred in multiple discrete steps, with initial KGF gene duplication and dispersion taking place in gibbon and involving loci corresponding to human chromosomes 15 and 21. These findings support the concept of a closer evolutionary relationship of human and chimpanzee and a possible selective pressure for such dispersion during the evolution of higher primates.
Keratinocyte growth factor (KGF, FGF7) is a member of the heparin-binding fibroblast growth factor family with a distinctive target cell specificity. While it is produced by cells of mesenchymal origin, it is active primarily, if not exclusively, on a wide variety of epithelial cell types (1, 2). A portion of the KGF gene, encoding its second and third exons, intervening intron, and 3′ untranslated region, was amplified in the evolution of higher primates and is present in multiple copies in the human genome (3). To better estimate the timing of these amplification events and explore the mechanisms responsible for dispersion of KGF-like sequences during primate evolution, we used a genomic clone containing one such amplified sequence to localize KGF-like genes in human and great ape chromosomes by fluorescence in situ hybridization (FISH). By this approach, which is uniquely suited for mapping multiple copy genes, we determined that KGF sequences are distributed to five chromosomes in human and chimpanzee, four in gorilla, two in orangutan, and one in gibbon. The identification and mapping of multiple loci of KGF-like sequences in human and great ape genomes provides insights in dating the amplification events and mechanisms involved as well as evidence for the phylogenetic relationships among greater apes and humans.
METHODS
KGF Genomic Clones and Primate Cells.
Approximately 330,000 colonies of a cosmid library (catalog no. 951202, Stratagene) were screened for sequences hybridizing to KGF. The library was constructed by ligation of partially MboI-digested human placental genomic DNA to the BamHI cloning site of pWE15. Colonies were lifted to nitrocellulose filters and hybridized with 32P end-labeled oligonucleotides K10 (exon 1, bases 446–494 of the KGF cDNA (2), K16 (exon 2, bases 732–790), and K20 (exon 3, bases 931–985)). Filters were washed with 0.1× standard saline citrate (SSC; 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.1% SDS at 50°C for 30 min. Autoradiography was performed and positive colonies were purified by subsequent rounds of screening. A single positive clone identified as p7–6A containing a 40-kb insert that hybridized at high stringency with KGF exon 2 and 3 sequences was identified. A genomic clone containing KGF promoter and exon 1 sequences (4) was the kind gift of P. Finch. Probes were hybridized to chromosomes derived from human peripheral lymphocytes of normal donors, and Epstein–Barr virus-transformed lymphoblastoid cell lines including CARL from chimpanzee (Pan troglodytes), ROC from gorilla (Gorilla gorilla), PUTI from orangutan (Pongo pygmaeus) and MLA 144 from gibbon (Hylobates lar) (American Type Culture Collection).
FISH.
The conditions for hybridization, detection of fluorescent signal, digital-image acquisition, processing, and analysis were carried out as described (5, 6). Duplicate sets of chromosome preparations were hybridized for high stringency post hybridization final washing at 60°C in 0.1× SSC, and low stringency wash at 37°C in 2× SSC. Chromosomes from human and great ape cells with specific hybridization signals were identified by rehybridization with centromeric or whole chromosome specific probes (Oncor and Vysis). The KGF signal was localized on G-banded chromosomes by trypsin or directly on LUT-inverted and contrast-enhanced digital images of 4′,6-diamidino-2-phenylindole-counterstained G-like banded chromosomes (7). The nomenclature and presumptive homologies of great ape chromosomes were according to the International System for Human Cytogenetics Nomenclature (8).
RESULTS
Localization of the KGF Gene to a Single Site on Human Chromosome 15.
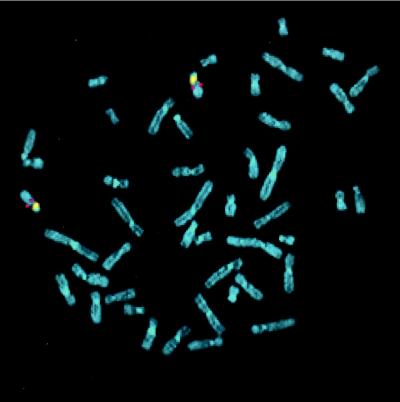
We initially sought to identify the unique chromosomal locus of the KGF gene in the human genome. Using a probe containing the KGF promoter region and exon 1 sequence, 226 of 260 labeled metaphases (86.92%) exhibited a double symmetrical fluorescent signal on both chromatids of chromosome 15 under low and high stringency posthybridization washing conditions (Fig. 1). Moreover, in the majority of the spreads, both chromosomes 15 were labeled as demonstrated by cohybridization with a chromosome 15-specific alpha satellite probe or G-banding (Fig. 1). The signal was localized in 50 well-banded chromosomes to region 15q15–21-1, which we assigned as the locus for the KGF gene (Fig. 2 a and b).
Figure 1.
Normal human chromosomes cohybridized with a digoxigenin-labeled cosmid probe for the exon 1 of KGF gene (red signal) and a biotinylated centromeric probe for chromosome 15 (yellow) counterstained with 4′,6-diaminido-2-phenylindole. Red rhodamine signal for KGF gene probe was observed on both chromosomes 15.
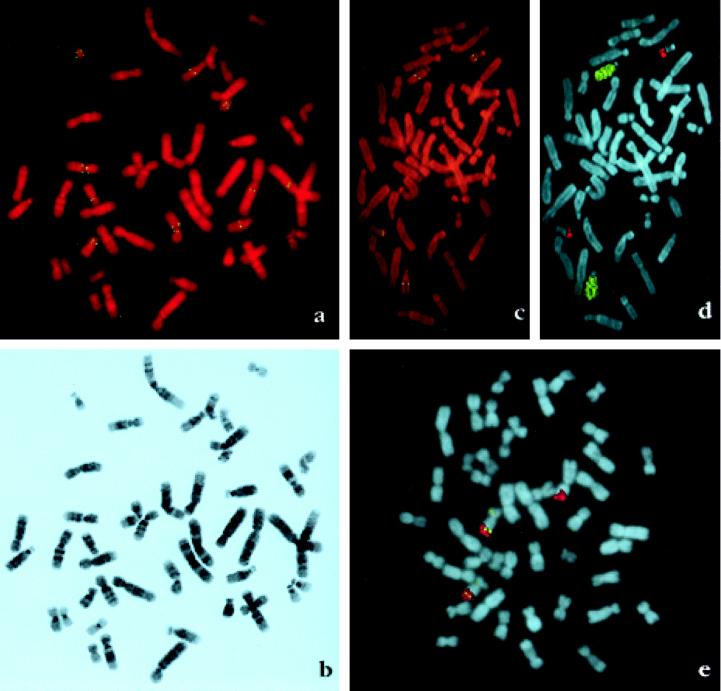
Figure 2.
(a) Digital images of normal human chromosomes hybridized with a biotinylated p7–6 cosmid probe comprising exons 2 and 3 counterstained with propidium iodide. Chromosomes 2, 9, 15, 18, and 21 exhibit double symmetrical fluorescent signals. Chromosomes 9 and 18 have signals on both short and long arms and chromosomes 21 have two pairs of signals on the long arm. (b) The same metaphase as in a showing G-like banding pattern generated by contrast enhancement and LUT (look-up table) inversion of the image of 4′,6-diamidino-2-phenylindole-restained chromosomes. The resolution of the chromosome bands permits an accurate individual chromosome identification as well as the regional localization of the fluorescent signal. (c) Orangutan chromosomes hybridized with a biotinylated p7–6 cosmid probe. Two chromosome pairs exhibit hybridization signal on the long arm. (d) The same metaphase as in c rehybridized with human chromosomes 15 (biotin-labeled, yellow) and 21 (digoxigenin-labeled, red) probes for chromosome identification. These probes painted the orangutan chromosomes homologous to human 15 and 21. The long arm of chromosomes 15 and the distal segment of the long arm of chromosome 22 where KGF sequences are located are painted. (e) Gibbon chromosomes hybridized with a biotinylated p7–6 probe (yellow signal) and a digoxigenin-labeled chromosome 21 probe. Chromosome 15 in gibbon has homology with human chromosomes 15 and 21. Two chromosomes 15 exhibit fluorescent signal on the distal part of the long arm corresponding to human chromosome 21 as shown by painting (red).
Localization of Amplified and Dispersed KGF-Like Sequences in the Human Genome.
Multiple copies of KGF exons 2 and 3 are present within human genomic DNA as determined by quantitative genomic hybridization techniques (3). By means of FISH, cosmid p7–6 A (containing KGF-like exon 2 and 3 sequences) was found to hybridize specifically to regions on chromosomes 2, 9, 15, 18, and 21 under standard hybridization conditions, and low and high stringency posthybridization washing. The majority of the 200 randomly selected metaphases had five pairs of homologous chromosomes with a double fluorescent signal. However, occasionally the signal was observed in only one homolog. A representative metaphase with signals on all five chromosome pairs is shown in Fig. 2a. A total of eight copies of KGF exon 2 and 3 per haploid genome was detected, with chromosomes 9, 18 and 21 each having two distinct sites of KGF-like sequences. The location of the fluorescent signal was determined on 25 banded chromosome pairs and placed on a G-band idiogram of ≈400 bands (International System for Human Cytogenetic Nomenclature 1985) (Fig. 2b). By this approach, we assigned KGF-like sequences to regions 2q21, 9p11, 9q12, 15q15, 18p11, 18q11, 21q11, and 21q21 (Fig. 2 a and b).
Localization of Amplified KGF-Like Sequences During Great Ape Radiation.
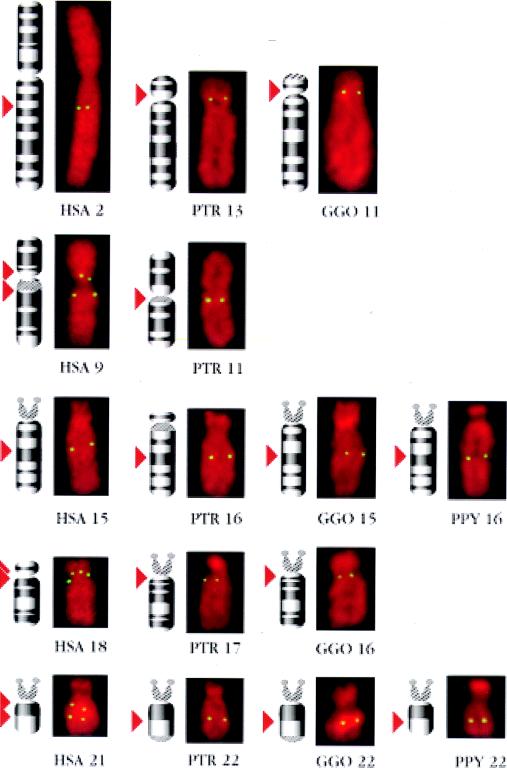
Multiple copies of KGF-like genes were detected by Southern blot analysis in gorilla and chimpanzee but only a single copy in orangutan, lesser apes, Old World monkey, mouse, and chicken, suggesting that amplification of this gene occurred during the evolution of higher primates (3). FISH analysis of chromosomes derived from great apes using the p7–6A cosmid probe showed that chimpanzee, gorilla orangutan and gibbon (Fig. 2 c and d) possessed five, four, two, or one locus (loci), respectively, recognized by the p7–6A probe (Fig. 2e). In duplicate hybridization experiments using low and high stringency posthybridization washing conditions, ≈300 labeled metaphases were recorded. Subsequently the same preparations were rehybridized with one or two of the human probes specific for chromosomes 2, 9, 15, 18, and 21, known to hybridize to the homologous chromosomes in great apes. In all four great ape species, KGF-like sequences were localized to chromosomes homologous to those containing KGF-like sequences in the human. KGF-like sequences were localized on chromosome 15q in gibbon; 16q15–21 and 22q22 in orangutan; 11p11, 15q15–21, 16q11–12, and 22q22 in gorilla; and 11q11, 13p11, 16q14–21, 17q11–12, and 22q21 in chimpanzee (Fig. 3). Chromosome 15 in gibbon has homology with human chromosome 15 and 21 (9). In 200 chromosome spreads examined, 180 had KGF-like sequences on the distal segment of the long arm corresponding with human chromosome 21, close to the interface of the regions homologous to human chromosomes 15 and 21 (Fig. 2e). In five spreads signal was also observed on the short arm of one chromosome 15. Thus, FISH data with KGF exon 2 and 3 suggest that gene duplication and dispersion occurred in a stepwise manner during the course of primate evolution, and began prior to the divergence of great apes and man.
Figure 3.
Comparable sized digital images of human (HSA), chimpanzee (PTR), gorilla (GGO), and orangutan (PPY) single chromosomes showing hybridization fluorescent signals with corresponding idiograms on their left side. The position of KGF hybridization signal on the idiograms is indicated by red arrows.
DISCUSSION
Our present investigations utilizing FISH analysis of KGF gene and KGF-related sequences have made it possible to track the radiational dispersion of this gene during primate evolution. In human, the KGF gene mapped to region 15q15–21.1, consistent with somatic cell hybrid analysis which indicated that the KGF gene was present on chromosome 15 (3). At least seven other copies of KGF-like sequences per haploid genome were localized on chromosomes 2, 9, 18, and 21. These additional KGF-like sequences cannot be attributed to duplication of entire chromosomes because such events occurred hundreds of millions of years ago (10–12), not in the relatively recent evolutionary timeframe in which amplification of KGF-like sequences has occurred. Portions of other genes, such as phosphoglycerate kinase (13), have been duplicated through a mechanism involving reverse transcription that produces sequences lacking introns. However, the presence of functional introns in the KGF-like sequences within the human genome indicates that the other mechanism(s) must be involved in their amplification (3).
KGF-like sequences (including the KGF gene) were localized to five, four, and two sites in chimpanzee, gorilla, and orangutan, respectively, with each site on a separate chromosome. These results establish a minimum number of copies of KGF-like sequences in these organisms. The present data are consistent with earlier evidence that KGF sequences were more numerous in human, chimpanzee, and gorilla as compared with orangutan. A single KGF copy number in orangutan was previously estimated both by slot-blot hybridization and restriction analysis (3). Thus, analysis by FISH is a more sensitive means to evaluate the multiplicity of recently amplified genes such as KGF, where nucleotide sequence divergence in the region around the gene may be minimal. We further established by FISH the existence of only a single chromosomal locus in gibbon. Therefore, it would appear that KGF gene duplication began at least 13–16 million years ago, prior to the divergence of orangutan from the human-chimpanzee-gorilla clad.
The localization of distinct chromosomal sites containing KGF-like sequences in human and great ape species suggests that their amplification occurred in a stepwise manner during evolution. Based on the present analysis, the initial KGF gene duplication event and dispersion involved loci in regions corresponding to human chromosomes 15 and 21. Surprisingly, the KGF locus on gibbon chromosome 15 mapped to an area homologous to human chromosome 21 rather than 15, where the human KGF gene is located. This observation suggested that the original site of the full-length KGF gene might have been in an area corresponding to human chromosome 21 instead of 15. However, considering the high frequency of rearrangements in the gibbon genome (9), it is possible that the KGF locus determined in this species was the result of a rearrangement. Similarly, the low incidence (5/200 spreads) of a second hybridization signal on gibbon chromosome 15 may be indicative of a rearrangement.
During primate evolution, additional KGF-like sequences were distributed to chromosomes corresponding to human chromosomes 2 and 18. This pattern is common to human, chimpanzee, and gorilla and, therefore, likely occurred prior to divergence of these species from their common ancestor ≈5–8 million years ago. Another locus on human chromosome 9 is shared only with chimpanzee. It is highly unlikely that the dispersion of these sequences occurred as an early event, with subsequent selective losses in subhuman primates over time (14). Similarly, we believe it is unlikely that gene duplication and dispersion would occur in parallel at the same respective chromosomal sites in several primate species after they had diverged from a common ancestor. Moreover, it is also improbable that such changes would occur spontaneously in cell culture. Thus, the pattern of KGF amplification and dispersion provides strong evidence that human and chimpanzee evolved from a common ancestor subsequent to divergence of the line leading to gorilla. Further duplication events resulting in two loci on each of human chromosomes 9, 18, and 21 are unique to the human and thus, presumably represent the most recent developments in the expansion and dispersion of KGF-like sequences. Analysis of these additional loci may advance our understanding of molecular events associated with human speciation.
The KGF-like sequence detected on the long arm of human chromosome 2 also provides new evidence for a previously described ancestral chromosomal fusion that distinguishes the human genome from that of the great apes. Telomeric fusion of chromosome 11 (gorilla) or 13 (chimpanzee) with chromosome 12 accounts for the reduction of 24 pairs in great apes to 23 pairs in humans (9, 15). The fusion of ancestral chromosomes occurred at human band 2q13 and resulted in loss of heterochromatic distal segments accompanied or followed by inactivation or elimination of one of the ancestral centromeres (15, 16). Hybridization experiments demonstrated remnant ancestral alphoid centromere sequences at band 2q21 (17). We observed that a KGF-like sequence is localized to the same band. In chimpanzee and gorilla, a KGF-like sequence was identified near the centromere at bands 13p11 and 11p11, which respectively correspond to human band 2q21. Thus, the KGF-like sequence at this band site is another marker indicating the contribution of band 13p11/11p11 to the formation of human chromosome 2.
The remarkable propensity of KGF-like sequences to become amplified and dispersed among several chromosomes raises the possibility that specific sequences within or near these loci may have conferred a predisposition for such processes to occur. Alphoid DNA, highly repetitive short (≈170 bp) DNA segments arranged in tandem arrays, has been implicated in genomic rearrangements (18) and may be relevant in the present context. All five human chromosomes containing KGF-like sequences belong to the same superchromosomal family, SF2, defined on the basis of similar alphoid DNA content (18, 19). The probability of seven new copies of KGF-like sequences being limited to the 11 chromosomes of SF2, assuming an equal probability of insertion into each chromosome, is (11/23)7 or 0.0057. Because the 11 chromosomes in SF2 comprise ≈40% of the human genome (20), a similar analysis based on chromosomal length would yield a probability of (0.4)7 = 0.0016. Thus, there is a highly significant correlation between the distribution of KGF-like sequences and alphoid DNA.
The association of KGF-like sequences and alphoid DNA is reinforced by their common, predominantly pericentromeric location. Moreover, in two instances when KGF-like loci were found outside pericentromeric regions (2q21 and 9q13), alphoid DNA was identified at the same sites (17). Interestingly, the probe used to detect alphoid DNA at these loci contained the 17-bp sequence element bound by centromere protein B. This DNA segment, a component of certain alphoid DNA sequences, was recently shown to be present in orangutan, gorilla, chimpanzee, and human DNA but absent from gibbon (21). Therefore, KGF-like sequences and a subset of alphoid DNA may share a common evolutionary history during the radiation of higher primates. Nucleotide sequence analysis of genomic fragments containing KGF-like sequences and investigation of their potential alphoid DNA content may provide insights into the mechanisms of duplication and dispersion responsible for the propagation of KGF-like sequences.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: KGF, keratinocyte growth factor; FISH, fluorescence in situ hybridization.
References
- 1.Rubin J S, Osada H, Finch P W, Taylor W G, Rudikoff S, Aaronson S A. Proc Natl Acad Sci USA. 1989;86:802–806. doi: 10.1073/pnas.86.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finch P W, Rubin J S, Miki T, Ron D, Aaronson S A. Science. 1989;245:752–755. doi: 10.1126/science.2475908. [DOI] [PubMed] [Google Scholar]
- 3.Kelley M J, Pech M, Suanez H N, Rubin J S, O’Brien S J, Aaronson S A. Proc Natl Acad Sci USA. 1992;89:9287–9291. doi: 10.1073/pnas.89.19.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finch P W, Lengel C, Chedid M. J Biol Chem. 1995;270:11230–11237. doi: 10.1074/jbc.270.19.11230. [DOI] [PubMed] [Google Scholar]
- 5.Popescu N, Zimonjic D, Hatch C, Bonner W. Genomics. 1994;20:333–335. doi: 10.1006/geno.1994.1182. [DOI] [PubMed] [Google Scholar]
- 6.Zimonjic D B, Popescu N C, Matsui T, Ito M, Chihara K. Cytogenet Cell Genet. 1994;65:184–185. doi: 10.1159/000133628. [DOI] [PubMed] [Google Scholar]
- 7.Zimonjic D B, Rezanka L, DiPaolo J A, Popescu N C. Cancer Genet Cytogenet. 1995;80:100–102. doi: 10.1016/0165-4608(94)00161-4. [DOI] [PubMed] [Google Scholar]
- 8.Harnden D G, Klinger H P, editors. An International System for Human Cytogenetic Nomenclature. Basel; in collaboration with Cytogenet. Cell Genet.: Karger; 1985. [Google Scholar]
- 9.Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T. Proc Natl Acad Sci USA. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno S, Wolf U, Atkin N B. Hereditas. 1968;59:169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x. [DOI] [PubMed] [Google Scholar]
- 11.Ohno S. Sex Chromosomes and Sex Linked Genes. New York: Springer; 1967. [Google Scholar]
- 12.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 13.McCarry J R, Thomas K. Nature (London) 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- 14.Minghetti P P, Dugaiczyk A. Proc Natl Acad Sci USA. 1993;90:1872–1876. doi: 10.1073/pnas.90.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ijdo J W, Baldini A, Ward D C, Reeders S T, Wells R A. Proc Natl Acad Sci USA. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luke S, Verma R S. Nat Genet. 1992;2:11–12. doi: 10.1038/ng0992-11. [DOI] [PubMed] [Google Scholar]
- 17.Baldini A, Ried T, Shridhar V, Ogura K, D’Aiuto, Rocchi M, Ward D C. Hum Genet. 1993;90:577–583. doi: 10.1007/BF00202474. [DOI] [PubMed] [Google Scholar]
- 18.Alexandrov I A, Mitkevich S P, Yurov Y B. Chromosoma. 1988;96:443–453. doi: 10.1007/BF00303039. [DOI] [PubMed] [Google Scholar]
- 19.Alexandrov I A, Mashkova T D, Akopian T A, Medvedev L I, Kisselev L L, Mitkevich S P, Yurov Y B. Genomics. 1991;11:15–23. doi: 10.1016/0888-7543(91)90097-x. [DOI] [PubMed] [Google Scholar]
- 20.Heslop-Harrison J S, Leitch A R, Schwarzacher T, Smith J B, Atkinson M D, Bennett M D. Hum Genet. 1989;84:27–34. doi: 10.1007/BF00210666. [DOI] [PubMed] [Google Scholar]
- 21.Haaf T, Mater A G, Wienberg J, Ward D C. J Mol Evol. 1995;41:487–491. doi: 10.1007/BF00160320. [DOI] [PubMed] [Google Scholar]