Abstract
The murine gene CHD1 (MmCHD1) was previously isolated in a search for proteins that bound a DNA promoter element. The presence of chromo (chromatin organization modifier) domains and an SNF2-related helicase/ATPase domain led to speculation that this gene regulated chromatin structure or gene transcription. This study describes the cloning and characterization of three novel human genes related to MmCHD1. Examination of sequence databases produced several more related genes, most of which were not known to be similar to MmCHD1, yielding a total of 12 highly conserved CHD genes from organisms as diverse as yeast and mammals. The major region of sequence variation is in the C-terminal part of the protein, a region with DNA-binding activity in MmCHD1. Targeted deletion of ScCHD1, the sole Saccharomyces cerevesiae CHD gene, was performed with deletion strains being less sensitive than wild type to the cytotoxic effect of 6-azauracil. This finding suggested that enhanced transcriptional arrest at RNA polymerase II pause sites due to 6-azauracil-induced nucleotide pool depletion was reduced in the deletion strain and that ScCHD1 inhibited transcription. This observation, along with the known roles of other proteins with chromo or SNF2-related helicase/ATPase domains, suggests that alteration of gene expression by CHD genes might occur by modifications of chromatin structure, with altered access of the transcriptional apparatus to its chromosomal DNA template.
Attempting to clone κY, a DNA-binding nuclear protein with affinity for Ig promoters, Delmas et al. (1) identified a novel murine protein with chromo (chromatin organization modifier) domains, SNF2-related helicase/ATPase domain and a DNA-binding domain (GenBank accession no. L10410). To denote the presence of these domains the protein was named CHD1. For clarity, it will be referred to as MmCHD1.
Chromo domains are found in a variety of proteins including the Drosophila melanogaster proteins HP1, which binds to heterochromatin at chromocenters and telomeres, and Polycomb, an inhibitor of homeotic gene expression responsible for maintaining a repressive heterochromatic structure at loci on polytene chromosomes (2). Although the mechanism by which chromo domains interact with heterochromatin is unclear, synthetic chromo domain peptides self-associate, giving chromo domain-containing proteins the potential to bind to each other and form complexes with as yet undetermined components of heterochromatin (3).
The SNF2-related family of proteins each contain a region of ≈400 amino acids with seven highly conserved helicase motifs (4). There are several subfamilies of SNF2-related genes, but none have chromo domains or DNA-binding domains similar to those found in MmCHD1. Some act either as activators (e.g., SNF2 and Brahma) or inhibitors (e.g., MOT1) of transcription whereas others are involved in DNA repair (e.g., RAD16 and ERCC6) or recombination (e.g., RAD54). Although none of the proteins coded for by these genes have been shown to possess helicase activity, several are DNA-dependent ATPases activated by single-stranded, double-stranded, or nucleosomal DNA. Mutations in SNF2 are suppressed by reductions in the levels of histones H2A or H2B, consistent with the suggestion that transcriptional activation occurs by remodeling of chromatin structure. It has been proposed that the basic function of the SNF2-related helicase/ATPase domain is to processively move along DNA templates and destabilize protein–DNA interactions (5).
The DNA-binding domain of MmCHD1 has been localized to a 229 aa region located near the C terminus of the protein (6). Binding to DNA occurred by minor groove interactions and was sequence-selective rather than sequence-specific, with a preference for (A+T)-rich tracts.
A number of other genes or gene fragments with similarity to MmCHD1 have since been cloned and sequenced. Relatively little is known about the interrelationships or functions of these genes. This study describes the identification and analysis of a group of genes with strong sequence similarity to MmCHD1, including three novel human genes. Comparative sequence analysis has allowed the identification of a highly conserved family of genes in eukaryotes ranging from yeast to human. Sequence motifs found in these genes suggest that they could be involved in a range of processes including DNA repair, regulation of transcription, and modification of chromatin structure. The identification of a highly conserved MmCHD1 homolog in Saccharomyces cerevesiae allowed the testing of hypotheses related to potential gene functions in an experimentally tractable system. Data are presented from these yeast experiments suggesting that CHD proteins inhibit transcription.
MATERIALS AND METHODS
Cloning and Sequencing.
PCR-amplified DNA probes were used for hybridization screening of a random/oligo(dT)-primed λZapII fetal brain cDNA library (Stratagene). Other clones were captured from a plasmid-based brain cDNA library with biotinylated oligonucleotides (Life Technologies). Clone coverage was extended using gene-specific primers and vector or adapter primers for PCR amplification of cDNA library or PCR-ready total fetus cDNA (CLONTECH). Expressed sequence tags (ESTs) for a D. melanogaster CHD gene were used to design primers to reverse transcription–PCR (RT-PCR) amplify intervening sequences from poly(A)+ RNA (CLONTECH). The cDNA clones were subsequently obtained for further sequencing (Berkeley Drosophila Genome Project, EST project). Human EST clones were obtained from the American Type Culture Collection.
Cycle sequencing reactions used double-stranded DNA as template. Fluorescently labeled dye terminators were incorporated into the reaction product with Amplitaq or Amplitaq FS (Perkin–Elmer/Applied Biosystems). Electrophoresis was carried out on models ABI373A and ABI377A sequencers (Applied Biosystems).
Sequence Analysis.
Primers for DNA sequencing and PCR were designed using the DNAStar (Madison, WI) primer select program. Resolution of ambiguous base calls and sequence contig assembly were performed with the DNAStar and sequencher programs. The blast algorithm (7) was used for database searches. Multiple sequence alignments were performed using clustal w version 1.6 with supplemental manual editing of gaps. Tree diagrams were constructed using the phylip Phylogeny Inference Package (8). protdist was used to calculate a distance matrix according to the Dayhoff PAM probability model (9), which was then used to estimate phylogenies by the Fitch–Margoliash least-squares distance method (10). All fitch runs were performed with global rearrangement and multiple jumbles (reordering of the data set 1,000 times) to evaluate the effect of different input orders on the derived trees and to assure that none of the subtrees had become caught in a statistical local minimum. consense was used to compute the consensus tree by the majority-rule method, and the final unrooted tree diagram was generated using drawgram.
Expression Analysis.
DNA probes corresponding to cDNA fragments near the 3′ ends of ORFs were amplified by PCR and labeled with [α-32P]dCTP prior to hybridization with Northern blots containing poly(A)+ RNA from a variety of human tissues (CLONTECH). Integrity of mRNA was tested with an actin cDNA probe after hybridization with the test probes.
Chromosomal Localization of Clones.
Sequence tagged sites for each of the human CHD genes were developed for PCR screening of DNA from the Genebridge4 radiation hybrid (RH) mapping panel or the Centre d’Étude du Polymorphisme Humain (CEPH) Mega-yeast artificial chromosome (YAC) library pools. Chromosomal localization was determined using the RHMapper algorithm or from a YAC-based physical map (both implemented at www-genome.wi.mit.edu).
Targeted Deletion of ScCHD1.
ScCHD1 was deleted by a modification of a PCR-based method (11). Chimeric primers containing 60 bp of sequence upstream or downstream of the ScCHD1 ORF and 20-bp flanking vector sequence were used to amplify a HIS3 marker from the pRS series of vectors (12). The resulting PCR product was transformed into the diploid strain YPH987 MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 leu2-Δ1/leu2-Δ1 his3-Δ200/his3-Δ200 CFIII(CEN3L.YPH983)TRP1SUP11. Replacement of ScCHD1 with the selectable marker HIS3 by homologous recombination was verified by PCR and Southern blot analyses. Tetrad dissection showed that the Scchd1Δ::HIS3 marker segregated 2:2 and that all four spores were viable. Strains YMB592 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 leu2-Δ1 his3-Δ200 Scchd1Δ1::HIS3) and its isogenic wild-type parent YMB590 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 leu2-Δ1 his3-Δ200) were used as indicated. Media for yeast growth and sporulation were as described (13), unless indicated otherwise.
Functional Analysis of ScCHD1.
Wild-type (YMB590), deletion (YMB592), and control strains were tested for several phenotypes. The effect of deletion of ScCHD1 on viability was determined by testing growth on yeast extract/peptone/dextrose (YPD) plates at 25°C, 30°C, or 37°C for 3 days. Sensitivity to UV radiation and hydroxyurea were determined as described (14). Methyl methanesulfonate (MMS) sensitivity was tested by growth on YPD plates with 0.01% MMS at 25°C for 2–3 days. Effects on mating efficiency were determined by standard mating assays (15). Telomeric silencing experiments were performed as described (16). Resistance to 6-azauracil (6AU, Sigma, catalogue no. A-1757) was tested in strains YMB592 and its isogenic wild-type strain YMB669 (YMB590 containing pRS313) transformed with either a HindIII–PvuII DNA fragment containing ScCHD1 cloned into pRS425 or vector only. Strains were grown to early logarithmic phase in SC-His-Leu medium at 30°C, harvested, washed, and dilutions with ≈200 cells were plated in triplicate on SC-His-Leu plates with 0–1250 μg/ml 6AU. The number of colony-forming units was determined after 4–5 days at 30°C.
RESULTS
Identification, Cloning, and Sequencing of Human Homologs of MmCHD1.
During the generation of a YAC contig on chromosome 15q26 an end-clone from CEPH YAC 748h5 was produced and sequenced. Database searches showed that a portion of this clone was closely related to MmCHD1. The complete MmCHD1 nucleotide sequence was used to query the EST subset of GenBank (release 84.0, August 1994) and four human EST sequences with strong similarities to MmCHD1 were identified (GenBank accession nos. M85593, T05810, T05827, and T06785). The cDNA clones from which these ESTs were derived were obtained and sequenced. Sequence comparisons indicated that the cDNA and YAC end clones were derived from three distinct genes. Further library screening, RT-PCR, and 5′-RACE (rapid amplification of cDNA ends) were used to generate three distinct human cDNA clone contigs 5,947 (GenBank AF006513), 7,764 (AF006514) and 6,331 bp (AF006515) long.
Sequence Analysis.
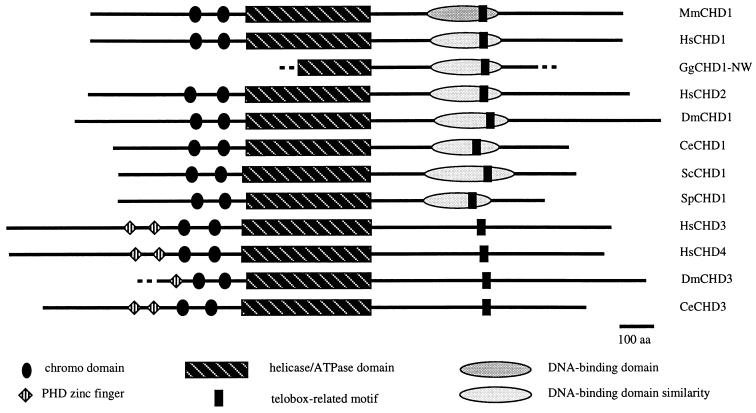
Translation from the first in-frame ATG in a sequence context consistent with translation initiation to the first in-frame stop codon predicted the existence of peptides of 1,709, 1,739, and 1,944 aa that were named HsCHD1, HsCHD2, and HsCHD3, based on their degree of similarity to the 1,711-aa MmCHD1. Like MmCHD1, each of the human CHD proteins was characterized by regions of reduced amino acid sequence complexity at their N and C termini. Similarly, the human proteins also had paired chromo domains near their N termini and centrally placed SNF2-related helicase/ATPase domains. Unlike the other proteins, HsCHD3 had two N-terminal zinc fingers of the PHD type (17). The psort algorithm (18) identified bipartite nuclear localization signals consistent with nuclear expression in all four CHD genes.
Protein sequences for MmCHD1, HsCHD1, HsCHD2, and HsCHD3 were aligned to allow comparison of sequence features, selections from which are shown in Fig. 1. The extent of amino acid identity and similarity between the three human proteins and MmCHD1 within the functional domains and over the entire coding regions are indicated in Table 1. The most highly conserved regions were within the helicase/ATPase domains. Even outside this domain, HsCHD1 had a very high degree of overall amino acid sequence conservation with MmCHD1 and most likely represents the human ortholog of the murine gene. Within that part of MmCHD1 possessing DNA-binding activity there is almost 60% identity between MmCHD1 and HsCHD2 suggesting the possibility of conservation of function. There is only limited similarity between MmCHD1 and HsCHD3 in this part of the protein.
Figure 1.
Amino acid alignments of selected regions of cloned human CHD proteins with MmCHD1. Identical residues are boxed in black and conserved residues shaded gray. Numbers represent residue positions within each sequence. Panels indicate regions of the alignment showing (a) chromo domains and selected segments of the (b) helicase/ATPase and (c) DNA-binding domains.
Expression Analysis.
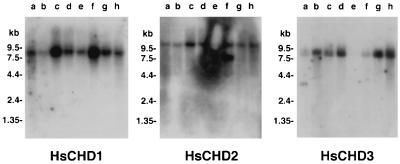
Taking advantage of the relatively low degree of sequence similarity at their 3′ ends, locus-specific DNA probes from these regions were prepared by PCR and hybridized to Northern blots. Fig. 2 shows the presence of transcripts of ≈8.5, 11, and 8 kb for HsCHD1, HsCHD2, and HsCHD3, respectively. Levels of expression varied substantially between different tissues. In the case of HsCHD2 an extra 7.5-kb band as well as the 11-kb band was noted in skeletal muscle, indicating the presence of either a tissue-specific splice variant or the use of an alternative polyadenylation signal. Both messages could code for the complete 5.2-kb HsCHD2 ORF.
Figure 2.
Northern blots for HsCHD1, HsCHD2, and HsCHD3. Lanes contain 2 μg poly(A)+ RNA extracted from heart (lane a), brain (lane b), placenta (lane c), lung (lane d), liver (lane e), skeletal muscle (lane f), kidney (lane g), and pancreas (lane h).
Chromosomal Localization of the HsCHD Genes.
HsCHD2 was independently mapped to 15q26 by PCR screening of the Genebridge4 RH mapping panel, lying between markers WI-6813 and D15S157. Similarly, HsCHD3 mapped to 17p13 between WI-9178 and D17S786. Owing to an inability to create a PCR assay for HsCHD1 that could distinguish between human and rodent loci it was not possible to map this gene with the RH panel. Instead, the CEPH YAC library was screened by PCR with clones 904b1 and 815c5 being positive. These YACs belong to a contig in 5q15-21 containing the marker WI-5811.
Delineation of the CHD Gene Family.
The mammalian CHD gene sequences were used to query public sequence databases. Three closely related genes were identified from large-scale genome sequencing efforts. Although no homolog was found in prokaryotes, the complete S. cerevesiae genomic sequence contained a single budding yeast gene with both chromo and helicase/ATPase domains (GenBank accession no. L10718), here termed ScCHD1. The blastp smallest sum probability score between HsCHD1 and ScCHD1 was 1.8 × 10−217. The continuing Caenorhabditis elegans large-scale sequencing project has produced two genomic DNA sequences predicted to code for CHD proteins, CeCHD1 (cosmid H06O01, J. Sulston, Cambridge, U.K.) and CeCHD3 (GenBank accession no. Z67884). Names for these genes were based on their similarity to other CHD genes (see below).
A number of related sequences were identified by more ad hoc cloning and sequencing efforts, although only one was initially recognized as coding for a member of the CHD gene family, this being a D. melanogaster homolog of MmCHD1, referred to here as DmCHD1 (GenBank accession no. X99021) (19).
A cDNA clone (GenBank accession no. D14316) very closely related to MmCHD1 and HsCHD1 was identified in a screen for proteins binding to the chicken δ1-crystallin gene enhancer (20). More recent work has shown there to be two, apparently near identical, avian CHD1 homologs (21, 22). One of these, GgCHD1-W, mapped to the W chromosome (the avian heterogametic sex chromosome), the other, GgCHD1-NW, which corresponded to the cDNA clone described above, was not W-linked.
Cloned components of the nuclear autoantigen Mi-2 also belonged to the CHD gene family. Antibodies to Mi-2 are found specifically in patients with dermatomyositis (23). Expression clones reacting with Mi-2 antisera coded for a peptide (GenBank accession no. U08379) that, apart from a frame-shift error near the 5′ end of the sequence and a 12-bp insertion also absent from an RT-PCR product amplified from total fetus RNA, was identical to a fragment of HsCHD3 (24). Seelig et al. (25) identified a full-length protein also recognized by anti-Mi-2 sera (GenBank accession no. X86691) that was very similar, but not identical to, HsCHD3. It may be confusing to identify either of these distinct sequences as Mi-2 and they will be referred to here as HsCHD3 and HsCHD4, respectively.
In addition to the full-length DmCHD1 gene described above, ESTs more closely related to HsCHD3 and HsCHD4 were found from two D. melanogaster cDNA clones (GenBank accession nos. AA140664, AA1640665, and AA202037). RT-PCR from D. melanogaster poly(A)+ RNA was used to span gaps between the ESTs. This DNA was cloned and, with the subsequently obtained cDNA clones, used to assemble a 4,911-bp cDNA contig which coded for a C-terminal fragment of DmCHD3 (GenBank accession no. AF007780).
Multiple ESTs were found for probable murine orthologs of the four human CHD genes. Although full-length sequences are not available for murine CHD genes except MmCHD1, from the mouse EST sequences that were available, other murine CHD genes also appear to be very similar to their human counterparts. No human or mouse ESTs (dbEST release 05/09/97) were identified for other, as yet uncharacterized, CHD genes.
In addition to the above-mentioned genes, a GenBank entry (X99021) represented an unannotated Schizosaccharomyces pombe CHD gene (SpCHD1).
Sequence Features and Phylogenetic Analysis of the CHD Gene Family.
The relative locations of functional motifs and domains found in the CHD family of proteins are depicted in Fig. 3. Each of the genes for which complete sequence is available shows consistent placement and spacing of two chromo domains toward the N termini and a central helicase/ATPase domain. All genes had nuclear localization signals and are predicted to have nuclear expression by psort (18). A subset of the genes, forming the CHD3 subfamily, also contain paired N-terminal PHD zinc fingers (except DmCHD3, for which complete sequence is not available). Multiple sequence alignments were performed using predicted amino acid sequences for each member of the CHD gene family, and the results of alignments are freely available on the National Human Genome Research Institute web site (http://www.nhgri.nih.gov/DIR/LGT/CHD). In addition to the protein motifs discussed above, analysis revealed the presence of a region with similarity to the Myb–DNA-binding domain. ScCHD1 contains an exact match to the consensus pattern for signature 1 of this motif (prosite pattern PS00037) with greatest similarity to the telobox subset of Myb-related DNA-binding motifs (26). The majority of the remaining CHD genes have 24-aa segments matching most of the residues that define the N-terminal portion of the telobox consensus sequence. The CHD3 subfamily has limited similarity to the telobox motif, but some residues are conserved.
Figure 3.
Protein domains found within CHD gene family members. Gene names are as described in the text. Extent of available sequence is indicated for GgCHD1-NW and DmCHD3; all other sequences are full length.
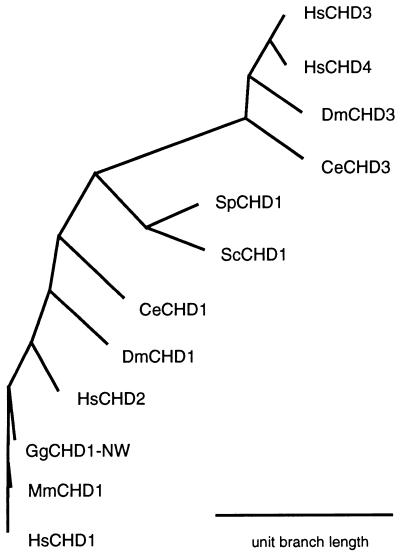
Clustering analysis based on the portion of the multiple sequence alignment for which all of the CHD genes were represented was used to determine phylogenetic relationships between each of the genes. Graphical representation of this analysis (Fig. 4) confirms impressions that HsCHD3, HsCHD4, DmCHD3, and CeCHD3 form a distinct subfamily of CHD proteins.
Figure 4.
Clustering relationships between members of the CHD family. Multiple sequence alignment of region for which overlapping sequences were available were used with the phylip Phylogeny Inference Package to generate unrooted fitch trees. drawgram was used to plot the tree diagram. The scale bar indicates map units per unit branch length, as determined by fitch.
Studies of CHD Gene Function in S. cerevesiae.
Deletion of ScCHD1 showed that it was not essential for haploid growth at 25°C, 30°C, or 37°C. Knowledge of the functions of other genes with chromo or SNF2-related helicase/ATPase domains prompted the examination of possible roles for ScCHD1 in DNA repair or effects on chromatin structure or transcription.
Deletion of ScCHD1 in a haploid yeast strain (YMB592) did not result in increased sensitivity to 0.1 M hydroxyurea, 50–150 J/m2 UV radiation, or 0.01% methyl methanesulfonate, as might be expected if this gene had a role in DNA repair. Similarly, in tests for the role of ScCHD1 in gene silencing in the vicinity of heterochromatin, the Scchd1Δ::HIS3 strain (YMB604) did not show detectable alteration of expression of marker genes compared with wild type in assays for telomeric position effects nor were any mating defects found compared with control strains.
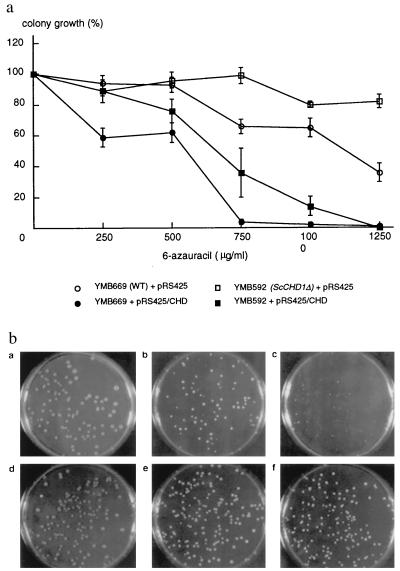
To test more generally for an effect of ScCHD1 on transcription, response to the pyrimidine analog 6AU was examined. The deletion strain (YMB592) was resistant to levels of the analog that reduced viability of wild-type cells (YMB669). Strain YMB592 showed ≈80% colony formation compared with 35% for strain YMB669 on plates containing 1250 μg/ml of 6AU (Fig. 5A). Overexpression of ScCHD1 increased sensitivity of the deletion and wild-type strains to 6AU with reduced viability even at 750 μg/ml of 6AU. Besides a quantitative difference in viability, differences in colony size between the two strains were also observed (Fig. 5B). In the presence of 500–1,000 μg/ml 6AU, the wild-type strain produced very small colonies that were heterogeneous in size compared with those of the deletion strain.
Figure 5.
(A) Effect of 6AU on yeast colony growth. Values for colony growth are normalized as a percent of untreated values. Error bars show standard error of the mean. Circles represent wild-type cells (YMB669) and squares strains in which ScCHD1 was deleted (YMB592). Filled symbols represent strains that were transformed with ScCHD1 cloned into pRS425 (pRS425/CHD), a high-copy number vector, whereas open symbols denote transformation with empty vector pRS425. (B) Appearance of yeast colonies after growth on yeast extract/peptone/dextrose (YPD) plates (a and d) or plates containing 500 μg/ml (b and e) or 1,000 μg/ml 6AU (c and f). Plates were spread with either wild-type (a, b, and c) or ScCHD1 deletion strains (d, e, and f).
DISCUSSION
This report establishes the CHD gene family as a group of highly conserved proteins sharing sequence motifs and functional domains associated with the regulation of chromatin structure and gene transcription. The CHD genes are unique in their combination of chromo, SNF2-related helicase/ATPase, and DNA-binding domains. It is of interest that the Drosophila gene polycomb, which has a chromo domain, acts as a suppressor of mutations in brm, a homeotic gene belonging to the SNF2 family. This finding suggests that these domains, which coordinately regulate chromatin structure and transcriptional activity through the action of separate molecules for polycomb–brm have had these activities integrated into a more tightly regulated control mechanism in the CHD genes.
Polyclonal antibodies raised against recombinant MmCHD1 have demonstrated an association with chromatin, with a fine granular staining pattern being seen in interphase nuclei (6). Staining was excluded from nucleoli and centromeric heterochromatin, and the protein was released from chromatin when chromosomes condensed during mitosis. Similarly, antisera from patients with dermatomyositis that recognized HsCHD3 and HsCHD4 also stained nuclei with a fine granular pattern with exclusion from the nucleoli (25). Further, affinity purified rabbit anti-Mi-2 antibodies immunoprecipitate several additional proteins from Hep-2 cell nuclear extracts suggesting that CHD genes may be part of multiprotein complexes.
Although there are several domains that have the potential to mediate such interactions, the components of chromatin to which CHD proteins bind are not clear. Chromo domains are required for targeting of Polycomb to its binding sites on polytene chromosomes in Drosophila. Polycomb itself has no DNA-binding activity and chromatin association occurs through protein–protein interactions (27). As has been described, a 229-aa segment near the C terminus of MmCHD1 binds (A+T)-rich DNA. Sequences within this region of the protein are not highly conserved throughout the CHD gene family, but each has a region similar to the telobox type of Myb-related DNA-binding domains (26). Although only found in the HsCHD3 subgroup of CHD proteins, PHD zinc fingers could also mediate an interaction with chromatin. The presence of a variety of protein motifs capable of associating with chromosomes allows the potential for targeting CHD gene family members to distinct regions or types of chromatin depending upon the specificity of motifs found in a particular CHD gene. Observed differences in patterns of tissue expression of HsCHD1, HsCHD2, and HsCHD3 might reflect such an effect. Antibodies to DmCHD1 have shown specific staining of only a subset of extended chromatin and puffed regions of polytene chromosomes associated with high transcriptional activity (19). It will be of great interest to determine whether DmCHD1 and DmCHD3 signals colocalize or are directed toward different chromosomal locations.
S. cerevesiae has a single CHD gene. This observation is in contrast to the situation in multicellular eukaryotes, with C. elegans and D. melanogaster each having a minimum of two, and humans and mice at least four CHD genes. Completing genomic sequences for other organisms will help to define details of the evolutionary process within the CHD gene family. Determination of the midpoint of the CHD gene phylogenetic tree using phylip (data not shown) shows that ScCHD1 and SpCHD1 are likely to be most representative of the archetypal CHD gene. The CHD3 subfamily appears as an outgroup on clustering analysis, and it will be interesting to determine whether any functional differences with other CHD genes are associated with the presence of such distinct features as the PHD zinc finger.
Strong sequence conservation between proteins from species at least a billion years removed from their most recent common ancestor implies strong conservation of function. In yeast, deletion of ScCHD1 was not found to have an effect on viability, growth under a variety of conditions, or response to DNA-damaging agents. The budding yeast genome contains a limited number of well-characterized regions of a heterochromatic nature (reviewed in ref. 28). In the current study, there was no evidence of altered silencing in the vicinity of the mating type or subtelomeric loci in a ScCHD1 deletion strain. A possible effect on chromatin structure at rDNA arrays (29) is under investigation.
As an assay of transcription, the response of ScCHD1 deficiency to 6AU was tested. After uptake into the cell, 6AU is converted to 6-azaUMP by uridine monophosphate (UMP) pyrophosphorylase. 6-azaUMP competitively inhibits orotidylic acid decarboxylase and inosine monophosphate (IMP) dehydrogenase, leading to reduced intracellular levels of GTP and UTP (30). Increased sensitivity to 6AU has been found in yeast mutants of RNA polymerase II and elongation factor SII. This effect is thought to be due to nucleotide pool depletion causing increased susceptibility to transcriptional arrest at RNA polymerase II pause sites (31). Resistance to 6AU in ScCHD1 deletion mutants thus could be due to loss of a transcriptional inhibitor. Resistance to 6AU has previously been noted for yeast mutants in UMP pyrophosphorylase or uracil uptake (32), but similar explanations are not likely to account for 6AU resistance in this case because of the probable nuclear localization of ScCHD1, the absence of any homology to metabolic enzymes or pore or transport proteins, and the observation that overexpression of ScCHD1 not only reversed 6AU resistance but resulted in 6AU hypersensitivity. It is not clear whether the proposed inhibitory effect of ScCHD1 on gene expression is general or, like SNF2, limited to the control of expression of a relatively small number of critical targets.
Although as yet there is no definitive evidence pointing to specific modes of action of the CHD genes, it is possible to propose a general model in which they function as negative regulators of gene expression by modifying extended, transcriptionally active chromatin to a more heterochromatic form, limiting access of the transcriptional apparatus to chromosomal DNA. In higher organisms, varying combinations of protein domains with affinity for DNA or chromatin may serve to direct CHD genes, or complexes containing CHD genes, toward distinct locations on decondensed chromosomes producing differing specificities for particular CHD genes. Preliminary experiments suggest that expression of portions of ScCHD1 from which the C-terminal putative DNA-binding domain have been deleted are not able to restore 6AU sensitivity to the deletion strain. If other CHD genes are able to functionally complement reduced 6AU sensitivity, it may be possible to define the specificity of different DNA-binding domains and examine associated alterations in gene expression and chromatin structure.
Table 1.
Amino acid sequence identities and similarities (%) of human CHD genes compared with MmCHD1
| HsCHD1 | HsCHD2 | HsCHD3 | |
|---|---|---|---|
| Chromo domains | |||
| Identity | 96.1 | 63.6 | 36.4 |
| Similarity | 98.7 | 77.9 | 51.9 |
| Helicase/ATPase domain | |||
| Identity | 99.5 | 86.8 | 54.0 |
| Similarity | 99.5 | 92.3 | 68.8 |
| DNA-binding domain | |||
| Identity | 97.4 | 59.4 | 9.2 |
| Similarity | 99.1 | 75.1 | 17.9 |
| Entire gene | |||
| Identity | 95.5 | 58.6 | 22.9 |
| Similarity | 97.4 | 69.5 | 34.5 |
Calculations based on chromo domains only (40 + 37 aa), helicase/ATPase domains (400 aa), and DNA-binding domains (229 aa).
Acknowledgments
We thank C. Robbins and N. Dietrich for their assistance with DNA sequencing. G. Hartzog generously provided a plasmid containing ScCHD1, and C. Baker Brachmann and D. Morrow made gifts of yeast strains. They, along with L. Pillus, M. Kenna, and other members of the Hieter/Boeke and Collins labs, gave valuable advice.
ABBREVIATIONS
- 6AU
6-azauracil
- EST
expressed sequence tag
- RH
radiation hybrid
- RT-PCR
reverse transcription–PCR
- YAC
yeast artificial chromosome
- CEPH
Centre d’Étude du Polymorphisme Humain
Footnotes
References
- 1.Delmas V, Stokes D G, Perry R P. Proc Natl Acad Sci USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh P B, Miller J R, Pearce J, Kothary R, Burton R D, Paro R, James T C, Gaunt S J. Nucleic Acids Res. 1991;19:789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowell I G, Austin C A. Biochim Biophys Acta. 1997;1337:198–206. doi: 10.1016/s0167-4838(96)00165-3. [DOI] [PubMed] [Google Scholar]
- 4.Eisen J A, Sweder K S, Hanawalt P C. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazin M J, Kadonaga J T. Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 6.Stokes D G, Perry R P. Mol Cell Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein J. phylip Phylogeny Inference Package. Seattle: Univ. of Washington; 1993. [Google Scholar]
- 9.Dayhoff M O. Atlas of Protein Sequence and Structure. Washington, DC: Natl. Biomed. Res. Found.; 1978. [Google Scholar]
- 10.Fitch W M, Margoliash E. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 11.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 14.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 15.Berkower C, Michaelis S. EMBO J. 1991;10:3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 17.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 18.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stokes D G, Tartof K D, Perry R P. Proc Natl Acad Sci USA. 1996;93:7137–7142. doi: 10.1073/pnas.93.14.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Development (Cambridge, UK) 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths R, Daan S, Dijkstra C. Proc R Soc London B. 1996;263:1251–1256. doi: 10.1098/rspb.1996.0184. [DOI] [PubMed] [Google Scholar]
- 22.Ellegren H. Proc R Soc London B. 1996;263:1635–1641. doi: 10.1098/rspb.1996.0239. [DOI] [PubMed] [Google Scholar]
- 23.Targoff I N, Reichlin M. Arthritis Rheum. 1985;28:796–803. doi: 10.1002/art.1780280711. [DOI] [PubMed] [Google Scholar]
- 24.Ge Q, Nilasena D S, O’Brien C A, Frank M B, Targoff I N. J Clin Invest. 1995;96:1730–1737. doi: 10.1172/JCI118218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seelig H P, Moosbrugger I, Ehrfeld H, Fink T, Renz M, Genth E. Arthritis Rheum. 1995;38:1389–1399. doi: 10.1002/art.1780381006. [DOI] [PubMed] [Google Scholar]
- 26.Bilaud T, Koering C E, Binet-Brasselet E, Ancelin K, Pollice A, Gasser S M, Gilson E. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller J. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 29.Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 30.Exinger F, Lacroute F. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 31.Archambault J, Lacroute F, Ruet A, Friesen J D. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jund R, Lacroute F. J Bacteriol. 1970;102:607–615. doi: 10.1128/jb.102.3.607-615.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]