Abstract
Extravascular procoagulant activity often accompanies cell-mediated immune responses and systemic administration of pharmacologic anticoagulants prevents cell-mediated delayed-type hypersensitivity reactions. These observations suggest a direct association between coagulation and cell-mediated immunity. The cytokine interleukin (IL)-4 potently suppresses cell-mediated immune responses, but its mechanism of action remains to be determined. Herein we demonstrate that the physiologic anticoagulant protein S is IL-4-inducible in primary T cells. Although protein S was known to inhibit the classic factor Va-dependent prothrombinase assembled by endothelial cells and platelets, we found that protein S also inhibits the factor Va-independent prothrombinase assembled by lymphoid cells. Thus, protein S-mediated down-regulation of lymphoid cell procoagulant activity may be one mechanism by which IL-4 antagonizes cell-mediated immunity.
Keywords: differential display, prothrombinase, cell-mediated immunity
Naive T cells differentiate into effector cells with distinct cytokine-producing capacities and functions after antigen-driven activation. Cell-mediated immunity is promoted by type I T helper (Th1) cells, which secrete interferon γ (IFN-γ), interleukin (IL) 2, and lymphotoxin, and humoral immunity is enhanced by type II T helper (Th2) cells, which secrete IL-4, IL-5, and IL-10. To effectively eradicate pathogens, the immune system must generate the appropriate type of Th cells. For example, inbred mouse strains that exhibit predominantly Th1 responses recover from infection with Leishmania major, but those that generate Th2 responses do not (1, 2). As IL-4 blocks the development of Th1 cells while promoting Th2 cell differentiation, the presence of IL-4 at the onset of an immune response can dictate whether cell-mediated or humoral immunity develops (for review, see ref. 3).
Although its physiologic significance is not understood, for some time it has been appreciated that a variety of cell-mediated inflammatory responses, including delayed-type hypersensitivity (4), experimental allergic encephalomyelitis (5), rheumatoid arthritis (6, 7), and allograft rejection (8), are associated with extravascular procoagulant activity. Moreover, delayed-type hypersensitivity responses can be prevented by systemic administration of pharmacologic anticoagulants (9–11), and allograft rejection correlates with decreased circulating levels of anticoagulant proteins (12). Although most clotting factors are generated by the liver, lymphoid cells can also produce components of the classical coagulation pathway and initiate the clotting process (for review, see refs. 13 and 14). Indeed, immunohistochemical studies have identified the procoagulants thrombin and factor X in lymphoid germinal centers (15) and interactions between T cells and monocytes/macrophage can induce procoagulant activity (16–22). This lymphoid-cell-associated procoagulant activity is up-regulated by the Th1 cytokine IFN-γ (23) and suppressed by the Th2 cytokines IL-4 and IL-10 (24–26). Thus, these findings strongly suggest that components of the coagulation pathway participate in cell-mediated immune responses.
The coagulant thrombin is synthesized and secreted as prothrombin, an inactive precursor. In the presence of a suitable cell surface containing negatively charged phospholipids, factor Xa assembles with factor Va to form a prothrombinase complex that coverts prothrombin to thrombin (for review, see ref. 27). In addition to the classic Xa/Va prothrombinase complex assembled by platelets and endothelial cells (28, 29), lymphoid cells also express novel prothrombinases, including one that is induced by viral infections (30) and another that incorporates effector cell protease receptor 1 (EPR-1), a factor Xa receptor that can substitute for factor Va in the prothrombinase complex (31). EPR-1 is constitutively expressed on monocytes and inducible on T cells after activation (32). Interestingly, an EPR-1-specific mAb potently suppresses cell-mediated immune responses (33), suggesting that prothrombinase activity may affect lymphocyte activity in vivo.
Physiologic suppression of the coagulation pathway is achieved by anticoagulant proteins that sterically and/or proteolytically inactivate the clotting factors. For example, protein S inhibits thrombin generation by inhibiting the activity of the classic Xa/Va prothrombinase (34–36) and by acting as a cofactor for activated protein C in the proteolytic inactivation of factors Va and VIIIa (37). In this report, we demonstrate that protein S is IL-4-inducible in primary T cells and that protein S inhibits the factor Va-independent lymphoid cell prothrombinase. Because procoagulant activity correlates with cell-mediated immunity, our findings suggest that IL-4-inducible expression of the anticoagulant protein S may be one mechanism by which this cytokine down-regulates cell-mediated responses.
MATERIALS AND METHODS
Differential Display (DD).
Murine T cells were purified from lymph nodes (LNs) of C57BL/6 mice (The Jackson Laboratory). B cells and macrophage were depleted via two rounds of complement treatment (Accurate Chemicals) using I-Ab-specific mAb (clone 28–16-8S). T cells (greater than 98% CD3+ by flow cytometry) were then isolated on Lympholyte M (Accurate Chemicals), washed, and frozen immediately or cultured for 24 hr on anti-CD3 precoated plates (clone 2C11, PharMingen) in the presence of IL-2 (50 units/ml) or IL-4 (500 units/ml). All cytokines were purchased from Genzyme. DD was performed essentially as described (38). Total RNA was prepared by using the guanidinium isothiocyanate method and reverse-transcribed with SuperScript II reverse transcriptase (GIBCO/BRL) using 1 μM “T-mer” as primer. DD reactions contained 20 ng of reverse-transcribed RNA, 1 μM of each primer, 1× PCR buffer (Boehringer Mannheim), all four dNTPs (each at 2 μM), 1 unit of Taq DNA polymerase (Boehringer Mannheim), and 1 μl of sequencing-grade 35S-labeled dATP (New England Nuclear/DuPont) in a final volume of 20 μl. Reactions were subjected to 40 cycles of 94°C for 30 sec, 40°C for 2 min, and 72°C for 30 sec, followed by a 10-min chase at 72°C in a Perkin–Elmer DNA thermal cycler. Reaction products were resolved on 6% denaturing polyacrylamide gels, dried without fixation, and subjected to autoradiography. The desired bands were excised from the gels and boiled in 50 μl of 10 mM Tris⋅HCl, pH 8/1 mM EDTA for 30 min. DNA was precipitated from the supernatants by using sodium acetate/ethanol in the presence of 50 μg of glycogen, and DD products in the precipitates were reamplified under the same PCR conditions described above, except that the volume and dNTP concentrations were increased to 50 μl and 200 μM, respectively. The reamplified DD products were resolved on preparative agarose gels and cloned (TA cloning kit, Invitrogen).
PCR.
Internal primers used for confirmation of the IL-4 inducibility of the DD product by PCR are indicated in Fig. 1c. Primers for the control hypoxanthine phosphoribosyltransferase gene were 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ and 5′-GATTCAACTTGCGCTCATCTTAGGC-3′, which generate a 162-bp PCR product. Reactions containing 1 μl of [32P]dCTP were subjected to 20 PCR cycles of 94°C for 30 sec, 55°C for 1 min, and 72°C for 30 sec and then resolved on 6% denaturing polyacrylamide gels. Products were detected by autoradiography. Control experiments established that conditions used were not saturating.
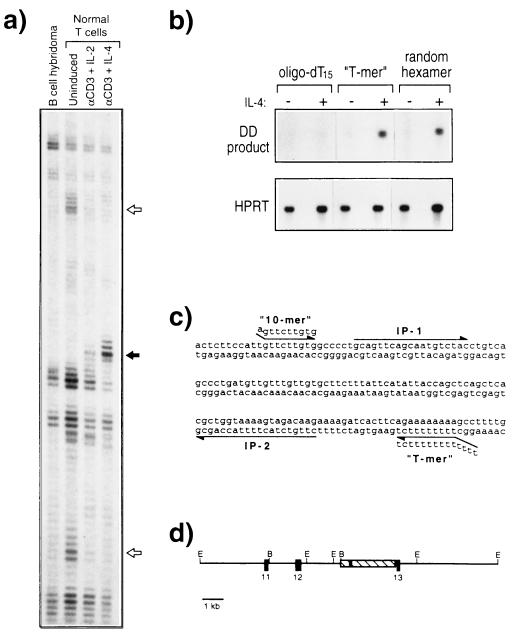
Figure 1.
Identification of protein S as an IL-4-inducible T cell gene. (a) DD of an IL-4-regulated gene fragment. The indicated RNA samples were subjected to DD by using the “10-mer” (5′-AGTTCTTGTG-3′) and “T-mer” (5′-T12CT-3′). Open arrows, two genes that are down-regulated upon T cell activation; solid arrow, the IL-4-inducible T cell gene characterized in this report. (b) PCR confirmation of the IL-4-inducible T cell gene. RNA was isolated from splenic cells cultured without cytokine (−) or in the presence of IL-4 at 500 units/ml (+) for 24 hr, and cDNA was prepared with (dT)15, the “T-mer” 5′-T12CT-3′, or random hexamers as indicated. PCR was conducted by using internal primers from the cloned DD product (Upper) or from the hypoxanthine phosphoribosyltransferase (HPRT) gene (Lower). (c) Sequence of the IL-4-inducible DD product. The sequence of the portion of a 3-kb clone encoding the DD product. The locations of the “10-mer” and “T-mer” used for DD and the internal primers (IP) used for PCR are indicated. (d) Localization of the DD product to an intron of the protein S gene. A restriction endonuclease enzyme map of the 16-kb genomic DNA fragment containing the DD product. The 3-kb clone isolated from a cDNA library is shown as a hatched rectangle, and the DD product is indicated by the solid region within this region. The locations of the cloned murine protein S exons homologous to the human protein S exons 11, 12, and 13 (46) are indicated. E, EcoRI; B, BamHI.
Libraries and Exon Trapping.
A murine IL-4-induced cDNA library was prepared by using the SuperScript choice system for cDNA synthesis (GIBCO/BRL) and polyadenylylated RNA isolated from splenic cells cultured with IL-4 (500 units/ml) for 24 hr and a mixture of (dT)15 and random hexamers. A strain 129 murine genomic library was provided by R. Jaenisch (The Whitehead Institute, Cambridge, MA). Exons were isolated by using the GIBCO/BRL exon trapping system. BamHI and EcoRI genomic fragments from the phage clone depicted in Fig. 1d were individually analyzed, and amplified exons were cloned (TA cloning kit, Invitrogen) and sequenced.
Northern Blot Analyses.
LN cells from C57BL/6, Stat6+/+, or Stat6−/− mice (39) were incubated in the presence of IL-2 (100 units/ml), IL-4 (500 units/ml, unless otherwise indicated), IL-12 (100 units/ml), or IFN-γ (500 units/ml). Where noted, plates were precoated with CD3-specific mAb 2C11 at 5 μg/ml. Peritoneal exudate lymphocytes (PELs) were generated by injection of 30 × 106 H-2b EL4 thymoma cells into the peritoneal cavity of BALB/c mice. Ten days later, peritoneal cells were harvested and host-derived major histocompatibility complex (MHC) class II I-A-positive cells and residual EL4 cells (MHC class I Kb/Db-positive) were removed by treatment with mAb (clones MKD6 and 28–8-6, respectively) followed by magnetic depletion (Advanced Magnetics, Cambridge, MA). CD8+ PELs were then isolated by treatment with mAb (clone 53.6) followed by magnetic enrichment. CD8+ LN cells were similarly isolated in parallel. The resultant populations were greater than 97% CD8+ CD3+ by flow cytometry. For Northern blots, 10 μg of total RNA was used per sample and ethidium bromide staining of agarose gels verified equivalent RNA loading. Blots were hybridized with a PCR-generated probe corresponding to nucleotides 1311–1991 of the protein S cDNA (40). The sizes of the detected protein S transcripts are consistent with previous reports (41).
Western Blotting.
LN cells from four C57BL/6 mice were harvested in RPMI 1640 medium containing 0.05% mouse serum. B cells were removed by two rounds of panning on sheep anti-mouse Ig precoated plates and the remaining cells were cultured for 24 hr on an anti-CD3 precoated plate in the presence of IL-4 (500 units/ml) in RPMI 1640 medium containing 0.05% mouse serum (IL-4 medium). Nonadherent cells (10 × 106, greater than 99% CD3+ by flow cytometry) were cultured in 0.5 ml of fresh IL-4 medium for 6 hr, at which point supernatant was collected for analysis. Purified human protein S standard was obtained from Enzyme Research Laboratories (South Bend, IN) and mouse plasma was obtained from Sigma. Samples were subjected to nonreducing SDS/PAGE using 7% acrylamide, transferred to nitrocellulose membrane (Schleicher & Schuell), and blocked with 4% milk in TBST-Ca (25 mM Tris/125 mM NaCl/0.5% Tween 20/2 mM CaCl2). Blots were then sequentially treated with affinity-purified goat anti-human protein S (34) at 2 μg/ml in 1% milk/TBST-Ca, biotinylated mouse anti-goat IgG (Pierce) at 1 μg/ml in 1% milk/TBST-Ca, a 1:2500 dilution of streptavidin-conjugated horseradish peroxidase (Kirkegaard & Perry Laboratories) in TBST-Ca, and enhanced chemiluminescence substrates (Bio-Rad).
Prothrombinase Assays.
Human peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood by using Ficoll/Hypaque (Pharmacia). To eliminate platelet contamination, cells were washed twice with 25 mM Hepes/135 mM NaCl/4 mM KCl/15 mM glucose, pH 7.4, twice with 25 mM Hepes/135 mM NaCl/4 mM KCl/5% BSA, pH 7.4, and twice with 25 mM Hepes/135 mM NaCl/4 mM KCl/15 mM glucose/3 mM CaCl2/BSA (3 mg/ml), pH 7.4 (complete buffer). PBMCs (2 × 106 cells) were then assayed for prothrombinase activity in 200 μl of complete buffer containing 100 pM human factor Xa (Enzyme Research Laboratories) and 14 nM human prothrombin (Enzyme Research Laboratories). Protein S (lot H133 S7) was purified by barium absorption of fresh frozen human plasma followed by immunoaffinity chromatography as described (35). Factor Xa was preincubated with protein S for 10 min at 37°C. PBMCs were then added for an additional 10 min, followed by prothrombin for 30 min, after which time the assay was terminated by the addition of 4 mM EDTA. Thrombin activity in supernatants was then quantitated with a commercially available enzymatic assay (ELCATECH, Winston-Salem, NC) and purified human α-thrombin (Enzyme Research Laboratories) as standard. For protein S neutralization studies, 2 μg of protein S was preincubated with 100 μg of protein S-specific mAb S4 or a control mouse IgG1 mAb in 50 μl for 45 min on ice and then assayed for its ability to inhibit prothrombinase activity as described above.
RESULTS
Protein S Is an IL-4-Inducible T Cell Gene.
We used DD (38) to identify genes specifically induced by IL-4 in T lymphocytes (Fig. 1a). Highly purified murine T cells were activated in vitro by using plate-bound CD3 mAb in the presence of either IL-2 or IL-4. Unstimulated T cells and a B cell hybridoma were analyzed in parallel, as controls. A 139-bp DD product dramatically induced by IL-4, but not IL-2, is shown in Fig. 1a. Northern blot analysis using this DD product as probe failed to reveal a hybridizing band, and no ORFs or significant homologies to other sequences in the genetic databases were identified. However, PCR analysis using internal primers confirmed that an IL-4-inducible gene fragment had been isolated (Fig. 1b). As (dT)15 was ineffective as a primer for cDNA preparation (Fig. 1b), we considered that an intronic sequence from a primary RNA transcript, rather than a spliced exonic sequence from a mature mRNA, may have been isolated. To evaluate this possibility, a genomic clone containing the DD sequence was obtained. Sequencing revealed that the DD product had likely arisen via priming of a short internal adenylate tract, rather than conventional priming of polyadenylylated RNA (Fig. 1c). The genomic clone was then subjected to an “exon-trapping” procedure (42), and three exons were isolated. Database searches of these exonic sequences revealed that they were identical to a contiguous segment of protein S cDNA (Fig. 1d). Inspection of the protein S promoter sequence revealed the presence of a consensus Stat6 binding site [TTCN4GAA (43, 44)] 558 nucleotides upstream of exon 1 (45), consistent with the IL-4 inducibility of this gene.
Protein S was first characterized as a vitamin K-dependent plasma-derived polypeptide (46) whose hereditary deficiency is a risk factor for venous thrombosis (47, 48). Like many factors of the coagulation pathway, synthesis of protein S is readily detected in hepatic tissue (49). However, protein S is also synthesized at extrahepatic sites, including the testis, ovary, brain, thymus, and spleen (49). In confirmation of the DD and PCR results, Northern blot analysis revealed that primary LN T cells activated in vitro in the presence of IL-4 express readily detectable levels of protein S transcripts (Fig. 2). The IL-4-mediated increase in protein S mRNA levels was specific, as protein S transcripts were not up-regulated in response to IL-2, IL-12, or IFN-γ, nor did these cytokines inhibit IL-4-mediated protein S induction (Fig. 2a). As is the case for other IL-4-regulated genes, IL-4-inducible protein S expression was Stat6-dependent (Fig. 2b).
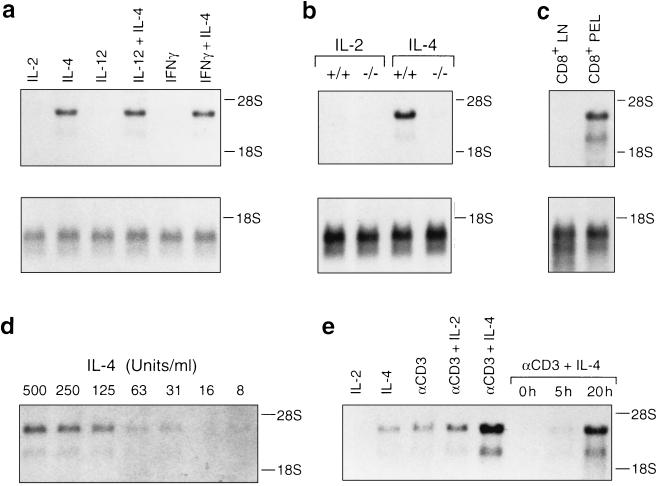
Figure 2.
Northern blot analysis of protein S expression in lymphocytes. (a) Protein S expression is specifically inducible by IL-4 and not inhibited by IL-12 or IFN-γ in LN T cells. (b) Protein S is IL-4-inducible in wild-type (+/+), but not Stat6-deficient (−/−) LN cells. (c) Protein S mRNA is expressed by freshly harvested CD8+ PELs but not CD8+ LN cells isolated in parallel. (d) Dose–response relationship for induction of protein S mRNA by IL-4. (e) Protein S is also inducible by T cell receptor-specific mAb (αCD3). Except for c, RNA was prepared from LN T cells treated with the indicated agents for 24 hr or for the time shown. Each lane contains 10 μg of total RNA. Northern blots were hybridized with a protein S probe. (a–c Lower) Reprobing of the same blots with a T cell receptor β-chain probe to confirm equal loading of RNA.
The IL-4-induced up-regulation of protein S mRNA levels was cell-type specific, as it was evident in both primary CD4+ and CD8+ T lymphocytes but not B lymphocytes (data not shown). Freshly harvested in vivo-activated CD8+ PELs, but not resting CD8+ lymphocytes, expressed high levels of protein S mRNA (Fig. 2c), indicating that activation-induced T cell expression of protein S was not an artifact of in vitro culture. As little IL-4 as 30 units/ml was sufficient to induce protein S expression (Fig. 2d). Protein S mRNA was also induced by T cell receptor stimulation, which led to synergistic increases in protein S expression in conjunction with IL-4 (Fig. 2e). Up-regulation of protein S mRNA was discernible within 5 hr and maximal by 20 hr after exposure to IL-4 (Fig. 2e and data not shown). In contrast to primary T cells, Th1 and Th2 T cell clones expressed only low or undetectable levels of protein S mRNA, independent of exposure to IL-4 (data not shown). These data demonstrate that protein S is an IL-4-inducible gene in primary T cells.
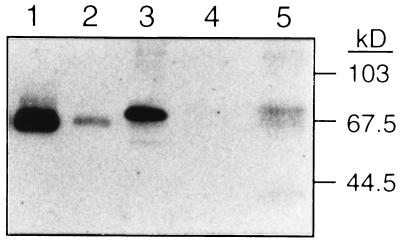
We next examined whether T cells secrete protein S polypeptide. Protein S secretion was evaluated by Western blotting using an affinity-purified goat anti-human protein S IgG that detects both human and murine protein S (Fig. 3, lanes 1–3). Supernatant was collected from highly purified (greater than 99% CD3+) murine T cells that had been activated by T cell receptor stimulation in the presence of IL-4. No immunoreactive material was detectable in the medium used for this experiment (lane 4). However, the T cell supernatant contained an immunoreactive band of the same apparent molecular weight as protein S in mouse plasma (lanes 5 and 3, respectively). Thus, primary murine T cells secrete protein S polypeptide upon activation in the presence of IL-4.
Figure 3.
Protein S is secreted by primary murine T cells. Western blot analysis of 500 pg of purified human protein S (lane 1), 50 pg of purified human protein S (lane 2), 0.1 μl of murine plasma (lane 3), 40 μl of control supernatant (lane 4), and 40 μl of supernatant from highly purified primary murine T cells (greater than 99% CD3+) activated in the presence of IL-4 (lane 5). The blot was developed with affinity-purified goat anti-human protein S (34).
Protein S Inhibits Lymphoid Cell Prothrombinase Activity.
The study of protein S-deficient individuals has established that protein S functions as a physiologic anticoagulant (47, 48). Mechanistically, protein S acts as a cofactor for activated protein C in the proteolytic degradation of factor Va (37) and also independently inhibits the factor Va/Xa prothrombinase complex (34–36). Unlike platelets and endothelial cells, lymphoid cells readily assemble prothrombinases in the absence of factor Va, because they express EPR-1, a cell-surface molecule that can substitute for factor Va (31, 32). Given that protein S was expressed by T cells, we examined its capacity to inhibit this factor Va-independent lymphoid procoagulant activity.
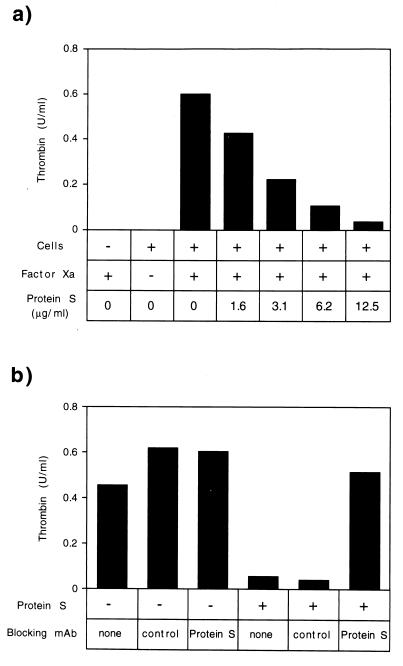
As shown in Fig. 4, when supplemented with only factor Xa, human PBMCs assembled a functional prothrombinase. PBMCs, factor Xa, and calcium ions were required to generate this activity (Fig. 4a and data not shown). Purified protein S inhibited this lymphoid cell prothrombinase, at concentrations similar to those previously shown to inhibit the factor Va-dependent prothrombinase on platelets and endothelial cells (Fig. 4a). A protein S-specific mAb blocked the ability of purified protein S to inhibit lymphoid cell prothrombinase activity (Fig. 4b). Thus, in addition to inhibiting the factor Va-dependent prothrombinase on platelets and endothelial cells (36), protein S also suppresses the factor Va-independent lymphoid cell prothrombinase.
Figure 4.
Protein S inhibits lymphoid cell prothrombinase activity. (a) Human PBMCs were isolated and prothrombinase activity was assayed in the presence or absence of human factor Xa and human protein S, as indicated. Control proteins BSA and protein C were inactive in this assay (data not shown). (b) The protein S mAb S4 specifically suppresses protein S-mediated antiprothrombinase activity. Protein S was used at 10 μg/ml.
DISCUSSION
In this report, we have shown that IL-4 promotes secretion of the anticoagulant protein S by T lymphocytes and that protein S inhibits lymphoid prothrombinase activity. Others have reported (50, 51) that protein S and another anticoagulant, activated protein C, suppress the production of tumor necrosis factor α (TNF-α) by monocytes. As TNF-α up-regulates monocyte procoagulant activity (23), IL-4-induced protein S secretion by T cells would antagonize both the expression and the function of lymphoid prothrombinase activity.
In vivo, IL-4 suppresses cell-mediated immunity. For example, inbred mouse strains that normally succumb to L. major. infections develop curing cell-mediated immune responses if endogenous IL-4 is neutralized (52). Because TNF-α is a critical component of cell-mediated immune responses (53), the capacity of protein S to suppress TNF-α production could be one means by which IL-4 antagonizes cell-mediated immunity. Thus, T cell-derived protein S may function as an IL-4-inducible antagonist of both prothrombinase activity and cell-mediated immune responses. Consistent with this hypothesis, mAb specific for the EPR-1 component of the lymphoid prothrombinase can suppress both prothrombinase activity (31, 32) and cell-mediated immunity (33).
The mechanisms by which some EPR-1-specific mAb antagonize cell-mediated immunity are unknown. It has been suggested that EPR-1 may function as a costimulatory molecule (33), perhaps transmitting signals upon binding factor Xa. In this model, the EPR-1-specific mAb suppresses lymphocyte activation by blocking EPR-1-mediated signals. Likewise, IL-4-inducible protein S might inhibit factor Xa/EPR-1 interactions, thus blocking EPR-1-dependent signaling. An alternative, but not mutually exclusive model, is that inhibition of prothrombinase activity, rather than inhibition of EPR-1-mediated signaling, is the mechanism by which EPR-1-specific mAb suppresses cell-mediated immunity. Although the epitope recognized by one of the immunosuppressive mAb is distinct from that required for factor Xa binding (54), it remains possible that this mAb may block prothrombinase activity indirectly, for example, by promoting EPR-1 endocytosis. This model would suggest that a downstream product of lymphoid prothrombinase activity promotes cell-mediated immune responses. Indeed, thrombin is known to be chemotactic for monocytes (55), mitogenic for lymphocytes (56), and proinflammatory in vivo (57). Moreover, in combination with submitogenic stimuli, thrombin costimulates T cell production of IL-2 (58). Thus, IL-4 might suppress cell-mediated immunity by inducing the expression of protein S, a prothrombinase antagonist.
Our studies suggest that the well-characterized highly conserved extrinsic coagulation pathway has evolved distinct functions in diverse tissues, including the immune system. Expression of protein S by neuronal (41), osteoid (59), and lymphoid cells had suggested functions beyond anticoagulation, but our findings suggest that the anticoagulant activity of protein S may be used for distinct outcomes in different tissues. Interestingly, activators of the monocyte fibrinolytic pathway are also IL-4-inducible (60), and fibrinolytic degradation products are immunosuppressive (61, 62). Further studies will be required to establish the precise physiologic roles of the coagulation and fibrinolytic pathways in the immune system. However, our findings suggest that local administration of pharmacologic agents that affect the balance between pro- and anticoagulants may permit antigen-specific manipulation of cell-mediated immune responses.
Acknowledgments
We thank Dr. Laurie Glimcher for advice and encouragement, Drs. Terri Laufer, Mark Kaplan, and Laurie Jackson-Grusby for helpful discussions and critical reading of the manuscript, and Yolanda Montejano for technical assistance with protein S purification. This work was supported by the Mathers Foundation and by National Institutes of Health Grants CA56462, AI40171 (M.J.G.), and HL21544 and HL52246 (J.H.G.).
ABBREVIATIONS
- IL
interleukin
- Th
T helper
- IFN-γ
interferon γ
- DD
differential display
- LN
lymph node
- PEL
peritoneal exudate lymphocyte
- PBMC
peripheral blood mononuclear cell
- EPR-1
effector cell protease receptor 1
- TNF
tumor necrosis factor
References
- 1.Heinzel F P, Sadick M D, Holaday B J, Coffman R L, Locksley R M. J Exp Med. 1989;169:59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinzel F P, Sadick M D, Mutha S S, Locksley R M. Proc Natl Acad Sci USA. 1991;88:7011–7015. doi: 10.1073/pnas.88.16.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiner S L, Seder R A. Curr Opin Immunol. 1995;7:360–366. doi: 10.1016/0952-7915(95)80111-1. [DOI] [PubMed] [Google Scholar]
- 4.Colvin R B, Johnson R A, Mihm M C, Jr, Dvorak H F. J Exp Med. 1973;138:686–698. doi: 10.1084/jem.138.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paterson P Y. Fed Proc. 1976;35:2428–2434. [PubMed] [Google Scholar]
- 6.Zacharski L R, Brown F E, Memoli V A, Kisiel W, Kudryk B J, Rousseau S M, Hunt J A, Dunwiddie C, Nutt E M. Clin Immunol Immunopathol. 1992;63:155–162. doi: 10.1016/0090-1229(92)90008-c. [DOI] [PubMed] [Google Scholar]
- 7.Gabazza E C, Osamu T, Yamakami T, Ibata H, Sato T, Sato Y, Shima T. Thromb Haemostasis. 1994;71:199–202. [PubMed] [Google Scholar]
- 8.Dvorak H F, Mihm M C, Jr, Dvorak A M, Barnes B A, Manseau E J, Galli S J. J Exp Med. 1979;150:322–337. doi: 10.1084/jem.150.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson D S. Immunology. 1965;9:219–234. [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S, Benacerraf B, McCluskey R T, Ovary Z. J Immunol. 1967;98:351–358. [PubMed] [Google Scholar]
- 11.Edwards R L, Rickles F R. Science. 1978;200:541–543. doi: 10.1126/science.644314. [DOI] [PubMed] [Google Scholar]
- 12.Tsuchida A, Salem H, Thomson N, Hancock W W. J Exp Med. 1992;175:81–90. doi: 10.1084/jem.175.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shands J W., Jr Haemostasis. 1984;14:373–377. doi: 10.1159/000215094. [DOI] [PubMed] [Google Scholar]
- 14.Edwards R L, Rickles F R. Semin Hematol. 1992;29:202–212. [PubMed] [Google Scholar]
- 15.Yamakawa M, Takagi M, Tajima K, Ohe S, Osanai T, Kudo S, Ito M, Sato T, Imai Y. Histochemistry. 1991;96:123–127. doi: 10.1007/BF00315982. [DOI] [PubMed] [Google Scholar]
- 16.Levy G A, Edgington T S. J Exp Med. 1980;151:1232–1244. doi: 10.1084/jem.151.5.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards R L, Rickles F R. J Immunol. 1980;125:606–609. [PubMed] [Google Scholar]
- 18.Geczy C L, Hopper K E. J Immunol. 1981;126:1059–1065. [PubMed] [Google Scholar]
- 19.Geczy C L, Meyer P A. J Immunol. 1982;128:331–336. [PubMed] [Google Scholar]
- 20.Helin H J, Fox R I, Edgington T S. J Immunol. 1983;131:749–752. [PubMed] [Google Scholar]
- 21.Geczy C L, Roberts I M, Meyer P, Bernard C C. J Immunol. 1984;133:3026–3036. [PubMed] [Google Scholar]
- 22.Geczy C, Raper R, Roberts I M, Meyer P, Bernard C C. J Neuroimmunol. 1985;9:179–191. doi: 10.1016/s0165-5728(85)80017-5. [DOI] [PubMed] [Google Scholar]
- 23.Schwager I, Jungi T W. Blood. 1994;83:152–160. [PubMed] [Google Scholar]
- 24.Herbert J M, Savi P, Laplace M C, Lale A. FEBS Lett. 1992;310:31–33. doi: 10.1016/0014-5793(92)81139-d. [DOI] [PubMed] [Google Scholar]
- 25.Pradier O, Gerard C, Delvaux A, Lybin M, Abramowicz D, Capel P, Velu T, Goldman M. Eur J Immunol. 1993;23:2700–2703. doi: 10.1002/eji.1830231048. [DOI] [PubMed] [Google Scholar]
- 26.Ramani M, Ollivier V, Ternisien C, Vu T, Elbim C, Hakim J, de Prost D. Br J Haematol. 1993;85:462–468. doi: 10.1111/j.1365-2141.1993.tb03333.x. [DOI] [PubMed] [Google Scholar]
- 27.Furie B, Furie B C. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 28.Tracy P B, Rohrbach M S, Mann K G. J Biol Chem. 1983;258:7264–7267. [PubMed] [Google Scholar]
- 29.Tracy P B, Eide L L, Mann K G. J Biol Chem. 1985;260:2119–2124. [PubMed] [Google Scholar]
- 30.Fung L S, Neil G, Leibowitz J, Cole E H, Chung S, Crow A, Levy G A. J Biol Chem. 1991;266:1789–1795. [PubMed] [Google Scholar]
- 31.Altieri D C, Edgington T S. J Biol Chem. 1989;264:2969–2972. [PubMed] [Google Scholar]
- 32.Altieri D C, Edgington T S. J Immunol. 1990;145:246–253. [PubMed] [Google Scholar]
- 33.Duchosal M A, Rothermel A L, McConahey P J, Dixon F J, Altieri D C. Nature (London) 1996;380:352–356. doi: 10.1038/380352a0. [DOI] [PubMed] [Google Scholar]
- 34.Heeb M J, Mesters R M, Tans G, Rosing J, Griffin J H. J Biol Chem. 1993;268:2872–2977. [PubMed] [Google Scholar]
- 35.Heeb M J, Rosing J, Bakker H M, Fernandez J A, Tans G, Griffin J H. Proc Natl Acad Sci USA. 1994;91:2728–2732. doi: 10.1073/pnas.91.7.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackeng T M, van’t Veer C, Meijers J C, Bouma B N. J Biol Chem. 1994;269:21051–21058. [PubMed] [Google Scholar]
- 37.Walker F J. J Biol Chem. 1980;255:5521–5524. [PubMed] [Google Scholar]
- 38.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 40.Chu M D, Sun J, Bird P. Biochim Biophys Acta. 1994;1217:325–328. doi: 10.1016/0167-4781(94)90294-1. [DOI] [PubMed] [Google Scholar]
- 41.Stitt T N, Conn G, Gore M, Lai C, Bruno J, Radziejewski C, Mattsson K, Fisher J, Gies D R, Jones P F, Masiakowski P, Ryan T E, Tobkes N J, Chen D H, DiStefano P S, Long G L, Basilicao C, Goldfarb M P, Lemke G, Glass D J, Yancopoulos G D. Cell. 1995;80:661–670. doi: 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- 42.Auch D, Reth M. Nucleic Acids Res. 1990;18:6743–6744. doi: 10.1093/nar/18.22.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotanides H, Reich N C. Science. 1993;262:1265–1267. doi: 10.1126/science.7694370. [DOI] [PubMed] [Google Scholar]
- 44.Schindler U, Wu P, Rothe M, Brasseur M, McKnight S L. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 45.Schmidel D K, Tatro A V, Phelps L G, Tomczak J A, Long G L. Biochemistry. 1990;29:7845–7852. doi: 10.1021/bi00486a010. [DOI] [PubMed] [Google Scholar]
- 46.Di Scipio R G, Hermodson M A, Yates S G, Davie E W. Biochemistry. 1977;16:698–706. doi: 10.1021/bi00623a022. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz H P, Fischer M, Hopmeier P, Batard M A, Griffin J H. Blood. 1984;64:1297–1300. [PubMed] [Google Scholar]
- 48.Comp P C, Esmon C T. N Engl J Med. 1984;311:1525–1528. doi: 10.1056/NEJM198412133112401. [DOI] [PubMed] [Google Scholar]
- 49.He X, Shen L, Bjartell A, Dahlback B. J Histochem Cytochem. 1995;43:85–96. doi: 10.1177/43.1.7822769. [DOI] [PubMed] [Google Scholar]
- 50.Hancock W W, Tsuchida A, Hau H, Thomson N M, Salem H H. Transplant Proc. 1992;24:2302–2303. [PubMed] [Google Scholar]
- 51.Grey S T, Tsuchida A, Hau H, Orthner C L, Salem H H, Hancock W W. J Immunol. 1994;153:3664–3672. [PubMed] [Google Scholar]
- 52.Sadick M D, Heinzel F P, Holaday B J, Pu R T, Dawkins R S, Locksley R M. J Exp Med. 1990;171:115–127. doi: 10.1084/jem.171.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vassalli P. Annu Rev Immunol. 1992;10:411–452. doi: 10.1146/annurev.iy.10.040192.002211. [DOI] [PubMed] [Google Scholar]
- 54.Ambrosini G, Altieri D C. J Biol Chem. 1996;271:1243–1248. doi: 10.1074/jbc.271.2.1243. [DOI] [PubMed] [Google Scholar]
- 55.Bar-Shavit R, Kahn A, Wilner G D, Fenton J W. Science. 1983;220:728–731. doi: 10.1126/science.6836310. [DOI] [PubMed] [Google Scholar]
- 56.Chen L B, Teng N N, Buchanan J M. Exp Cell Res. 1976;101:41–46. doi: 10.1016/0014-4827(76)90409-2. [DOI] [PubMed] [Google Scholar]
- 57.Cirino G, Cicala C, Bucci M R, Sorrentino L, Maraganore J M, Stone S R. J Exp Med. 1996;183:821–827. doi: 10.1084/jem.183.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mari B, Imbert V, Belhacene N, Far D F, Peyron J F, Pouyssegur J, Van Obberghen-Schilling E, Rossi B, Auberger P. J Biol Chem. 1994;269:8517–8523. [PubMed] [Google Scholar]
- 59.Maillard C, Berruyer M, Serre C M, Dechavanne M, Delmas P D. Endocrinology. 1992;130:1599–1604. doi: 10.1210/endo.130.3.1531628. [DOI] [PubMed] [Google Scholar]
- 60.Hart P H, Burgess D R, Vitti G F, Hamilton J A. Blood. 1989;74:1222–1225. [PubMed] [Google Scholar]
- 61.Girmann G, Pees H, Schwarze G, Scheurlen P G. Nature (London) 1976;259:399–401. doi: 10.1038/259399a0. [DOI] [PubMed] [Google Scholar]
- 62.Edgington T S, Curtiss L K, Plow E F. J Immunol. 1985;134:471–477. [PubMed] [Google Scholar]