Abstract
The adeno-associated virus 2 (AAV), a single-stranded DNA-containing, nonpathogenic human parvovirus, has gained attention as a potentially useful vector for human gene therapy. However, the single-stranded nature of the viral genome significantly impacts upon the transduction efficiency, because the second-strand viral DNA synthesis is the rate-limiting step. We hypothesized that a host-cell protein interacts with the single-stranded D sequence within the inverted terminal repeat structure of the AAV genome and prevents the viral second-strand DNA synthesis. Indeed, a cellular protein has been identified that interacts specifically and preferentially with the D sequence at the 3′ end of the AAV genome. This protein, designated the single-stranded D-sequence-binding protein (ssD-BP), is phosphorylated at tyrosine residues and blocks AAV-mediated transgene expression in infected cells by inhibiting the leading strand viral DNA synthesis. Inhibition of cellular protein tyrosine kinases by genistein results in dephosphorylation of the ssD-BP, leading not only to significant augmentation of transgene expression from recombinant AAV but also to autonomous replication of the wild-type AAV genome. Dephosphorylation of the ssD-BP also correlates with adenovirus infection, or expression of the adenovirus E4orf6 protein, which is known to induce AAV DNA replication and gene expression. Thus, phosphorylation state of the ssD-BP appears to play a crucial role in the life cycle of AAV and may prove to be an important determinant in the successful use of AAV-based vectors in human gene therapy.
Keywords: protein tyrosine kinases, DNA replication, gene expression, gene therapy
The adeno-associated virus 2 (AAV), a single-stranded DNA-containing parvovirus, so far has not been shown to be associated with any pathology in humans (1, 2). The wild-type (wt) AAV genome has been shown to integrate in a site-specific manner into human chromosome 19q13.3-qter (3–5). Thus, recombinant AAV vectors have emerged as useful alternatives to the more commonly used retroviral and adenoviral vectors for human gene therapy (6–10). However, recent studies from two independent laboratories have suggested that after infection, the leading strand viral DNA synthesis is a rate-limiting step in the efficient transduction by AAV vectors (11, 12). AAV inverted terminal repeats (ITRs) contain 145 nt each, the terminal 125 nt of which are palindromic and form T-shaped hairpin (HP) structures. The 3′-HP structure serves as a primer for AAV DNA replication. AAV ITRs also contain an additional domain, designated the D sequence, which is a stretch of 20 nt that is not involved in HP formation (1, 2, 13). Our previous studies have indicated that the D sequence plays a crucial role in the efficient rescue, selective replication, and encapsidation of the AAV genome (14, 15). We hypothesized that a cellular protein(s) might interact with the D sequence and prevent the second-strand viral DNA synthesis. Indeed, using electrophoretic mobility-shift assays (EMSAs), we have recently provided evidence for the existence of a hitherto unknown host-cell protein that specifically interacts with the double-stranded D sequence, which we have designated as the double-stranded D-sequence-binding protein (dsD-BP), and demonstrated that the dsD-BP is recruited by AAV to ensure efficient rescue and replication of the wt viral genome (15).
However, the AAV genome contains two D sequences, one in each ITR, that are single-stranded but complementary (13). The D(−) sequence is always at the 3′ end, whereas the D(+) sequence is invariably at the 5′ end. Although our recent studies have indicated that a cellular protein also interacts specifically with the single-stranded D sequence (16), it remains unclear whether the same dsD-BP also binds to single-stranded D sequences or, alternatively, there are two types of D-BP. What role the single-stranded D-sequence-binding protein (ssD-BP) plays in the AAV life cycle also remains unknown.
Previous studies have documented that coinfection with adenovirus, treatment with hydroxyurea (HU), or expression of the adenovirus E4orf6 protein leads to a significant enhancement in AAV transduction efficiency, because these treatments promote the second-strand viral DNA synthesis (11, 12, 17). To determine whether the ssD-BP is involved in the AAV second-strand DNA synthesis, we have carried out systematic analyses of the ssD-BP–D-sequence interactions. In this report, we present evidence that the ssD-BP forms a specific complex with the D sequence at the 3′ end of the AAV genome. This protein is phosphorylated at tyrosine residues and inhibits the leading strand viral DNA synthesis. Inhibition of cellular protein tyrosine kinases results in dephosphorylation of the ssD-BP, leading not only to significant augmentation of transgene expression of recombinant AAV but also autonomous replication of the wt AAV genome. Dephosphorylation of the ssD-BP also correlates with adenovirus infection or expression of the adenovirus E4orf6 protein, which is known to induce AAV DNA replication and gene expression. Thus, the phosphorylation state of the ssD-BP appears to play a crucial role in the life cycle of AAV.
MATERIALS AND METHODS
Cells, Viruses, and Plasmids.
Human cervical carcinoma cell line, HeLa, was obtained from the American Type Culture Collection and maintained as monolayer cultures in Iscove’s-modified Dulbecco’s medium supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics. Human AAV and adenovirus type 2 (Ad2) stocks were kindly supplied by Kenneth I. Berns (Cornell University Medical College, New York) and Kenneth H. Fife (Indiana University School of Medicine, Indianapolis), respectively. A recombinant plasmid, pCMV/E4orf6 (12), was generously provided by Richard J. Samulski (University of North Carolina, Chapel Hill).
Preparation of Whole Cell Extracts (WCEs).
WCEs were prepared according to the method of Muller (18) from HeLa cells under the following conditions: (i) cells were maintained either in 10% FBS or 0.5% FBS for 14 days or treated with 80 mM HU for 24 hr (17) before harvesting the cells; (ii) cells were either mock-infected or infected with the wt AAV at a multiplicity of infection (moi) of 1 or 10, infected with Ad2, or coinfected with wt AAV + Ad2 for 36 hr prior to harvesting the cells as previously described (14); (iii) cells were either mock-transfected or transfected separately with plasmids pWP-7A (19), pWP-19 (19), or pCMV/E4orf6 (12) using the lipofectamine reagent by the transfection protocol supplied by the vendor (GIBCO/BRL). Total protein concentration was determined by the Bio-Rad protein assay kit, and the extracts were frozen in liquid N2 and stored at −70°C.
EMSAs.
EMSAs were performed as described previously (14). Briefly, DNA-binding reactions were performed in a volume of 20 μl with 2 μg poly(dI-dC), 2 μg BSA, and 12% glycerol in Hepes buffer (pH 7.9). Ten micrograms of protein from each WCE was preincubated for 10 min at 25°C followed by the addition of 10,000 cpm of 32P-labeled D(−)-sequence synthetic oligonucleotide (5′-AGGAACCCCTAGTGATGGAG-3′) in the reaction mixture. The binding reaction was allowed to proceed for 30 min at 25°C, and the bound complexes were separated from the unbound probe on low-ionic strength 4% polyacrylamide gels with recirculating Tris-acetate-EDTA (TAE) buffer (pH 7.9) containing 6.72 mM Tris⋅HCl, 3.3 mM sodium acetate, and 1 mM EDTA. In some experiments, EMSAs were also carried out with 32P-labeled D(+)-sequence oligonucleotide (5′-CTCCATCACTAGGGGTTCCT-3′), or double-stranded D(±)-sequence, respectively, the latter of which was prepared by annealing the two complementary D(−) and D(+) sequences followed by purification on 10% polyacrylamide gels as previously described (14). In competition experiments, increasing concentrations of unlabeled D sequence, a non-AAV substitute sequence (S sequence) (5′-CCAATATTAGATCTGATATCA-3′) (14, 15), or the AAV-cap sequence (5′-ACCGTGGATACTAATGGCGTG-3′) were added to the reaction mixtures before the addition of the radiolabeled D sequence probe for 10 min; this was followed by incubation with the labeled probe as described above.
Immunoprecipitation.
Equivalent amounts (≈100 μg) of WCEs prepared from cells following mock-infection, infection with AAV or with Ad2, or treatment with HU were cleared of nonspecific binding by incubation with 20 μl of protein A agarose beads for 1 hr at 4°C in an orbital shaker. After preclearing, 2 μg of monoclonal anti-phosphotyrosine antibody (Upstate Biotechnology, Lake Placid, NY) was added and incubated at 4°C for 12 hr in the shaker followed by precipitation with protein A agarose beads as previously described (20–23). Supernatants were collected, and immunoprecipitates were washed three times with PBS and dissolved in 20 μl of EMSA reaction buffer at 25°C for 30 min. Ten microliters of each of the supernatants and resuspended pellet solutions were used for EMSA with the D(−) oligonucleotide probe as described above. In some experiments, immunoprecipitations were also carried out with monoclonal anti-p53 antibody (Upstate Biotechnology).
AAV DNA Replication Assays.
Approximately 5 × 105 HeLa cells were plated in 6-cm dishes for 12 hr followed by either no treatment or treatment with 80 mM HU for 24 hr, 150 μM genistein (24), or 1 mM sodium ortho-vanadate (NaOV) (22, 23) for 2 hr. Cells were then infected with the wt AAV at an moi of 2. Low-Mr DNA samples were isolated at various times postinfection by the procedure described by Hirt (25) and analyzed on Southern blots using a 32P-labeled AAV coding sequence-specific DNA probe as previously described (14, 15).
RESULTS
The D-BP Interacts Specifically and Preferentially with the Single-Stranded D Sequence in the AAV-ITR at the 3′ End of the Viral Genome.
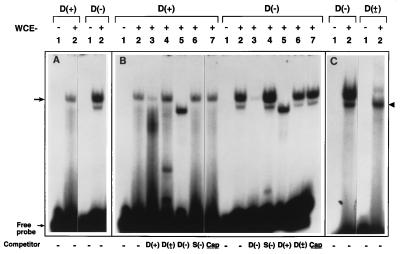
WCEs were prepared from HeLa cells, and EMSAs were carried out with each of the two single-stranded D sequence synthetic oligonucleotides, D(+) and D(−), under identical conditions (16). These results are shown in Fig. 1. The D(−) sequence was consistently and reproducibly at least 3-fold more efficient in forming a complex (arrow) with the D-BP compared with the D(+) sequence (Fig. 1A, lanes 2) as determined by densitometric scanning of autoradiographs. In some experiments, an additional faster-migrating complex with the D(−) sequence was also detected. Binding to each oligonucleotide was specific because it could be competed with the respective unlabeled oligonucleotides (Fig. 1B, lanes 3). The binding could not be competed with the double-stranded [D(±)] sequence, with a non-AAV, substitute S(−) sequence, or with an AAV-cap sequence (Fig. 1B, lanes 4, 6, and 7). Cross-competition experiments were also performed where binding to the D(−) oligonucleotide was competed with the unlabeled D(+) oligonucleotide and vice versa. Because these oligonucleotides are complementary, they can anneal to form the D(±) structure. Under these conditions, a DNA–protein complex was observed that migrated faster than that formed with either single-stranded oligonucleotide (Fig. 1B, lanes 5). To confirm that the faster-migrating complex did indeed consist of D-BP bound to the D(±) sequence, EMSAs were carried out with D(−) and D(±) oligonucleotides under identical conditions (Fig. 1C). Again, the D-BP interacting with the D(±) sequence migrated faster than that with the D(−) sequence (arrowhead). These results suggest that either the D-BP interacts preferentially with single-stranded rather than double-stranded D sequences, or two different D-BPs exist.
Figure 1.
Differential interaction of the D-BP with the D(−) and D(+) sequences in the AAV ITRs. EMSAs were carried out with D(−) or D(+) 20-nt synthetic oligonucleotides (A) as previously described (16). Various indicated unlabeled single- or double-stranded oligonucleotides representing the D sequence, a non-AAV substitute (S) sequence, and AAV capsid (cap) gene sequences were used in competition experiments (B). Comparative analysis of D-BP interaction with the single-stranded D(−) sequence versus the double-stranded D(±) sequence (C). The arrow indicates the single-stranded D-BP, and the arrowhead indicates the double-stranded D-BP.
The D-BP that Binds to Single-Stranded D Sequence Is Phosphorylated and Undergoes Dephosphorylation During Cell Cycle.
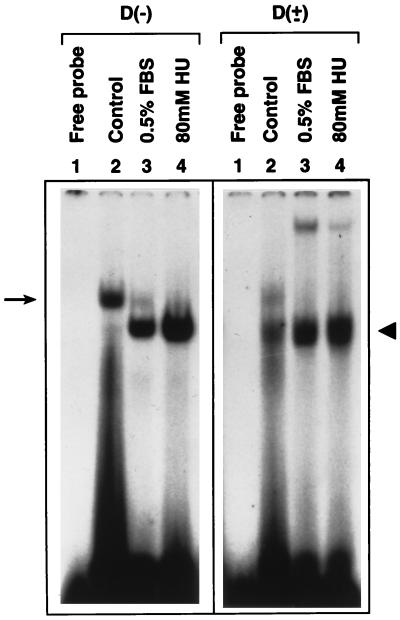
We next wished to determine whether binding of the D-BP to the D(−) sequence could be altered under conditions known to enhance AAV transduction efficiency, such as serum starvation and treatment with HU (12, 17). To this end, WCEs were prepared from HeLa cells that were maintained in 10% or 0.5% FBS for 14 days or treated with 80 mM HU for 24 hr and utilized in EMSAs with the D(−) oligonucleotide. As controls, the same WCEs were used in EMSAs with the D(±) oligonucleotide. These results are shown in Fig. 2. As observed before (Fig. 1), the D(−) probe interacted predominantly with the slower-migrating D-BP in WCEs prepared from untreated cells (Fig. 2, arrow). It is interesting to note, however, that the same D(−) sequence interacted with the faster-migrating D-BP in WCEs prepared from growth-inhibited cells. In contrast, there was no change in the migration pattern of the D-BP that interacted with the D(±) probe (arrowhead). These results suggest that the same D-BP that binds to single-stranded D sequence undergoes some type of modification during cell cycle, and there appear to be two forms of D-BP; one interacts with the single-stranded (ssD-BP) and the other interacts with the double-stranded (dsD-BP) D sequences.
Figure 2.
The ssD-BP undergoes modification following growth-arrest of HeLa cells. EMSAs were carried out with WCEs prepared under different conditions as described in the text. D(−) or D(±) probes were used under identical conditions as described in the legend to Fig. 1.
It was of interest to investigate next whether coinfection with Ad2 or expression of the AdE4orf6 protein, which recently was shown to be necessary and sufficient to catalyze the second- strand synthesis (12), also correlated with this modification. We hypothesized that among other possible posttranslational modifications, the ssD-BP from HeLa cells grown in 10% FBS that interacted with the D(−) probe was a phosphorylated protein and that various treatments led to dephosphorylation of the ssD-BP. This hypothesis was experimentally tested, as will be described later.
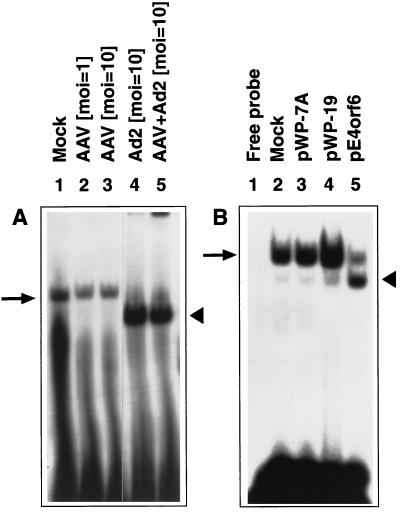
WCEs were prepared from mock-infected, AAV-infected, Ad2-infected, or AAV+Ad2 coinfected HeLa cells, as well as from cells that were mock-transfected, or transfected with plasmids pWP-7A (containing the TcR gene), pWP-19 (containing the neoR gene), or pCMV/E4orf6 (containing the AdE4orf6 gene), respectively. The results of EMSAs carried out with these WCEs and the D(−) probe are shown in Fig. 3. It is noteworthy that the D(−) sequence formed a complex with the phosphorylated form of the ssD-BP (Fig. 3A, arrow), and infection with AAV alone at an moi of 1 or 10 had no effect. The same probe formed another complex with the ssD-BP that was dephosphorylated in Ad2-infected or AAV + Ad2 coinfected cells (arrowhead). Remarkably, the AdE4orf6 protein alone, known to facilitate the viral second-strand DNA synthesis, was also sufficient to dephosphorylate the ssD-BP (Fig. 3B). Compared with near-total dephosphorylation of the ssD-BP following Ad2 infection, the incomplete conversion is most likely due to low transfection efficiency of pCMV/E4orf6 plasmid.
Figure 3.
Ad2 infection or the AdE4orf6 protein expression leads to ssD-BP modification. Cells were either mock-infected or infected with the wt AAV or Ad2, or were coinfected with AAV+Ad2 at the indicated moi, and WCEs prepared from these cells were used in EMSAs with the D(−) probe (A). EMSAs were also carried out with WCEs prepared from cells transfected with the indicated plasmids (B).
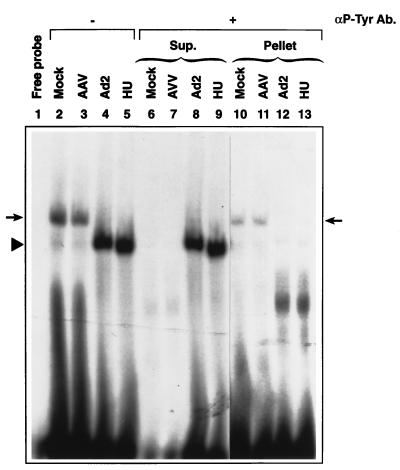
Using the anti-phosphotyrosine antibody, we have determined that the slow-migrating ssD-BP is indeed the phosphorylated protein that specifically interacts with the D(−) sequence, whereas the faster-migrating ssD-BP is dephosphorylated. These data are shown in Fig. 4. It is interesting to note that the complexes formed with WCEs prepared from mock-infected or AAV-infected cells (left arrow), but not from Ad-infected or HU-treated cells (arrowhead), could be immunoprecipitated with the anti-phosphotyrosine antibody. The phosphorylated ssD-BP could also be recovered from the immunoprecipitate and shown to interact with the D(−) sequence (right arrow).
Figure 4.
The ssD-BP is phosphorylated at tyrosine residues. The ssD-BP was immunoprecipitated from equivalent amounts of WCEs prepared from cells following mock-infection, infection with AAV or Ad2, or treatment with HU, with the anti-phosphotyrosine antibody (αP-Tyr Ab), followed by precipitation with protein A agarose beads as previously described (20–23). Supernatants were collected, and immunoprecipitates were washed with PBS and dissolved in EMSA reaction buffer. Ten microliters of each of the supernatants (Sup.) and resuspended pellet solution (Pellet) were used for EMSA with the D(−) oligonucleotide probe.
Inhibition of Phosphorylation of the ssD-BP Leads to Enhancement in the Transduction Efficiency of Recombinant AAV and Autonomous Replication of the Wt AAV Genome.
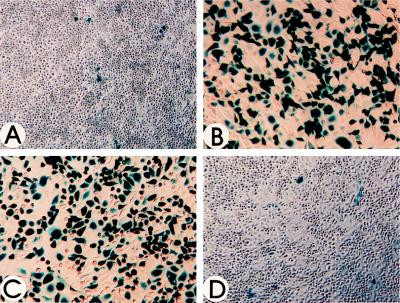
We next explored whether AAV transduction efficiency could be enhanced by simply inhibiting tyrosine phosphorylation of the ssD-BP. HeLa cells were treated for 2 hr with 150 μM genistein, a specific inhibitor of protein tyrosine kinases (24). The cells were then infected with a recombinant AAV vector containing the cytomegalovirus (CMV) promoter-driven lacZ gene (vCMVp-lacZ) at an moi of 20. After staining with X-Gal at 48 hr postinfection, the number of blue cells were enumerated as previously described (26, 27). As controls, cells were either mock-treated, treated with HU, or treated with NaOV, a known protein phosphatase inhibitor (22, 23), followed by infection with the recombinant AAV. These results are shown in Fig. 5. It is evident that whereas AAV transduction efficiency of HeLa cells after mock-treatment or treatment with NaOV was approximately 3%, the transduction efficiency increased to approximately 60% after treatment of HeLa cells with HU or genistein.
Figure 5.
Inhibition of protein tyrosine kinases leads to increased transduction efficiency of recombinant AAV. HeLa cells were infected with 20 moi of vCMVp-lacZ following either no treatment (A) or treatment with 10 mM HU for 24 hr (B), 150 μM genistein for 2 hr (C), or 1 mM NaOV for 2 hr (D). Forty-eight hours postinfection, cells were fixed and the recombinant AAV-mediated transgene expression was detected as previously described (26, 27).
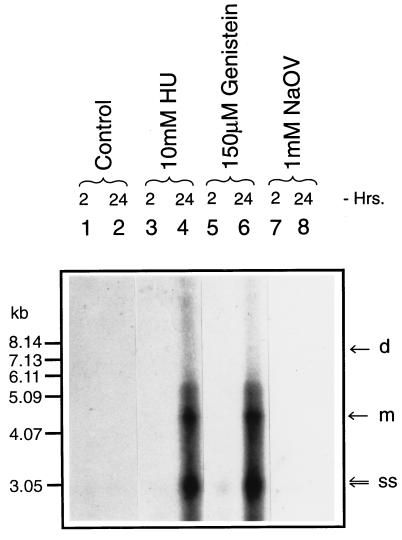
We also wished to examine whether dephosphorylation of the ssD-BP leads to autonomous replication of the wt AAV genome. HeLa cells were either mock-treated or treated with HU, genistein, or NaOV as described in Materials and Methods and infected with 2 moi of wt AAV. Low-Mr DNA samples were isolated and analyzed on Southern blots using an AAV-specific DNA probe (14, 15). It is evident that the inhibition of protein tyrosine kinases by genistein leads to autonomous replication of the wt AAV genome, the extent of which is comparable to that following treatment with HU (Fig. 6). These results strongly suggest that the phosphorylation state of the ssD-BP plays a crucial role in the life cycle of AAV.
Figure 6.
Inhibition of cellular protein tyrosine kinases leads to autonomous replication of the wt AAV genome. Cells were infected with the wt AAV at an moi of 2 following either no treatment or treatment with HU, genistein, or NaOV, and low-Mr DNA samples isolated at various indicated times postinfection were analyzed on Southern blots using an AAV-specific DNA probe. d, m, and ss denote the dimeric and monomeric replicative DNA intermediates and single-stranded progeny AAV genomes, respectively.
DISCUSSION
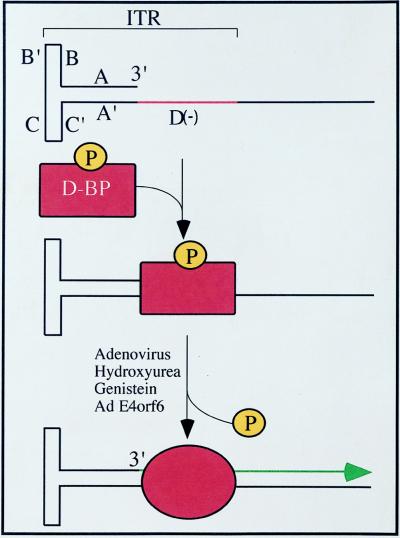
We have identified a previously unknown host-cell single-stranded DNA-binding protein, ssD-BP, which is phosphorylated at tyrosine residues. This protein is also phosphorylated at serine and threonine residues (data not shown). Interestingly, however, dephosphorylation of tyrosine residues in this protein appears to be necessary and sufficient to lead to second-strand DNA synthesis of the AAV genome in infected cells. At the very least, there is a very strong correlation between the dephosphorylation state of the ssD-BP and treatments known to provide a suitable environment for AAV replication and efficient transduction (K.Y.Q., B. Khuntirat, C. Mah, D.M.K., X.-S.W., S.P., S. Z. Zhou, V. Dwarki, M. C. Yoder and A.S., unpublished work). Based on the data presented, we propose a model for an inhibitory role of the phosphorylated form of the ssD-BP in AAV-mediated transgene expression. This model is shown in Fig. 7. In its phosphorylated form, the ssD-BP preferentially complexes with the D(−) sequence in the AAV ITR and blocks initiation of DNA replication from the 3′ OH end. Coinfection with Ad2, treatment with HU or genistein, or expression of the AdE4orf6 protein causes dephosphorylation of the ssD-BP, leading to some type of conformational change, which results in accessibility of the 3′ OH end as a primer for the second-strand DNA synthesis. It appears, therefore, that the host cell exerts a significant influence on the fate of the incoming single-stranded AAV genome and, to some extent, offers a plausible explanation why AAV-mediated transduction of different cell types varies widely (6–12). Indeed, our recent studies have documented a strong correlation between the dephosphorylation state of the ssD-BP and the efficiency of AAV-mediated transduction in vitro as well as in vivo (K.Y.Q., B. Khuntirat, C. Mah, D.M.K., X.-S.W., S.P., S. Z. Zhou, V. Dwarki, M. C. Yoder and A.S., unpublished work).
Figure 7.
A possible model for the role of the tyrosine-phosphorylated ssD-BP in the viral second-strand DNA synthesis and AAV-mediated transgene expression. The phosphorylated form of ssD-BP preferentially complexes with the D(−) sequence in the AAV ITR and blocks initiation of DNA replication from the 3′ OH end. Various indicated treatments cause dephosphorylation of the ssD-BP, leading to some type of conformational change, which results in accessibility of the 3′ OH end as a primer for the second-strand DNA synthesis followed by gene expression.
Although coinfection with adenovirus has long been known to provide the helper function for AAV replication and gene expression, the cellular target(s) that mediate that function has remained elusive. Similarly, treatment of cells with HU also allows wt AAV to undergo autonomous replication, but how an inhibitor of cellular DNA replication facilitates AAV replication has remained unclear. Both adenovirus coinfection and HU treatment have also been shown to enhance the transduction efficiency of recombinant AAV (11, 12, 17). Interestingly, both of these treatments also lead to dephosphorylation of the ssD-BP. Thus, a target host-cell single-stranded DNA-binding protein, which is phosphorylated at tyrosine residues, has been identified and appears to play a crucial role in AAV DNA replication and gene expression.
These observations are also consistent with the involvement of single-stranded DNA-binding proteins in prokaryotic and eukaryotic cellular DNA replication in general and in DNA replication of adenoviruses and herpesviruses in particular (28). Several cogent examples of specific interactions of cellular proteins with viral nucleic acids recently have been described. These include the identification of a site-specific DNA-binding protein that cooperates with the minute virus of mice nonstructural protein (NS1) to initiate the viral DNA replication (29) and the identification of a sequence-specific single-stranded DNA-binding protein, Pur α, which regulates DNA replication of the human neurotropic JC virus (30). Similarly, preferential interactions of host-cell proteins with 3′ ends of hepatitis A virus RNA (31) and hepatitis B virus pregenomic RNA (32) have also been observed. The AdE4orf6 protein-mediated transactivation of AAV transgene expression (12) coupled with the documented role of the AdE4orf6 protein in the interaction with the p53 tumor suppressor protein and p53-mediated transcriptional transactivation (33) thus may share similarities with intracellular events involving the D-sequence–ssD-BP interaction. In DNA–protein UV cross-linking experiments, we have determined the ssD-BP to be an approximately 53-kDa protein, but distinct from the p53 tumor suppressor protein, based on immunoprecipitation studies with monoclonal anti-p53 antibody (data not shown).
It is obvious that the ssD-BP plays an important role in the host cell. Indeed, a number of cellular target sequences in the human genome have been identified based on the D sequence homology search of DNA sequence databases (data not shown). It is clear, therefore, that further studies to identify the ssD-BP, and the availability of the cellular gene that encodes it, may be crucial for gaining an insight into the molecular steps in the AAV life cycle in general and in AAV-mediated gene transfer in particular.
Acknowledgments
We thank Dr. Richard J. Samulski for generously providing the pAdE4orf6 plasmid. We also thank Drs. Robert H. Schloemer and Mervin C. Yoder for a critical reading of this manuscript. This research was supported in part by Public Health Service Grants HL-48342, HL-53586, and DK-49218 (Centers of Excellence in Molecular Hematology) from the National Institutes of Health and by the Phi Beta Psi Sorority. A.S. was supported by an Established Investigator Award from the American Heart Association.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AAV, adeno-associated virus 2; ssD-BP and dsD-BP, single- and double-stranded D-sequence-binding protein, respectively; wt, wild type; ITR, inverted terminal repeat; EMSA, electrophoretic mobility-shift assay; WCE, whole cell extract; HU, hydroxyurea; moi, multiplicity of infection.
References
- 1.Berns K I, Bohenzky R A. Adv Virus Res. 1987;32:243–306. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 2.Berns K I, Giraud C. Curr Top Microbiol Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Proc Natl Acad Sci, USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotin R M, Menninger J C, Ward D C, Berns K I. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 5.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muzyczka N. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 7.Carter B J. Curr Opin Biotechnol. 1992;3:533–539. doi: 10.1016/0958-1669(92)90082-t. [DOI] [PubMed] [Google Scholar]
- 8.Flotte T R, Carter B J. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 9.Chatterjee S, Lu D, Podsakoff G, Wong K K., Jr Ann N Y Acad Sci. 1995;77:79–90. doi: 10.1111/j.1749-6632.1995.tb31045.x. [DOI] [PubMed] [Google Scholar]
- 10.Chatterjee S, Wong K K., Jr Curr Top Microbiol Immunol. 1996;218:61–73. doi: 10.1007/978-3-642-80207-2_5. [DOI] [PubMed] [Google Scholar]
- 11.Fisher K J, Gao G-P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari F K, Samulski T, Shenk T, Samulski R J. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava A, Lusby E W, Berns K I. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X-S, Ponnazhagan S, Srivastava A. J Mol Biol. 1995;250:573–580. doi: 10.1006/jmbi.1995.0398. [DOI] [PubMed] [Google Scholar]
- 15.Wang X-S, Ponnazhagan S, Srivastava A. J Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X-S, Qing K Y, Ponnazhagan S, Srivastava A. J Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell D W, Alexander I E, Miller A D. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller M T. J Virol. 1987;61:858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nahreini P, Woody M J, Zhou S Z, Srivastava A. Gene. 1993;124:257–262. doi: 10.1016/0378-1119(93)90402-o. [DOI] [PubMed] [Google Scholar]
- 20.Piwnica-Worms H, Saunders K B, Roberts T M, Smith A E, Cheng S H. Cell. 1987;49:75–82. doi: 10.1016/0092-8674(87)90757-4. [DOI] [PubMed] [Google Scholar]
- 21.Tokiwa G, Dikic I, Lev S, Schlessinger J. Science. 1996;273:792–794. doi: 10.1126/science.273.5276.792. [DOI] [PubMed] [Google Scholar]
- 22.Kumagai A, Dunphy W G. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 23.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Science. 1996;275:661–664. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 24.Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 25.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 26.Ponnazhagan S, Wang X S, Woody M J, Luo F, Kang L Y, Nallari M L, Munshi N C, Zhou S Z, Srivastava A. J Gen Virol. 1996;77:1111–1122. doi: 10.1099/0022-1317-77-6-1111. [DOI] [PubMed] [Google Scholar]
- 27.Ponnazhagan S, Mukherjee P, Yoder M C, Wang X S, Zhou S Z, Kaplan J, Wadsworth S, Srivastava A. Gene. 1997;190:203–210. doi: 10.1016/s0378-1119(96)00576-8. [DOI] [PubMed] [Google Scholar]
- 28.Williams K R, Chase J W. In: The Biology of Non-Specific DNA Protein Interactions. Rezvin A, editor. Boca Raton, FL: CRC; 1990. pp. 197–227. [Google Scholar]
- 29.Christensen J, Cotmore S F, Tattersall P. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C-F, Gallia G L, Muralidharan V, Chen N N, Zoltick P, Johnson E, Khalili K. J Virol. 1996;70:4150–4156. doi: 10.1128/jvi.70.6.4150-4156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusov Y, Weitz M, Dollenmeier G, Gauss-Muller V, Siegl G. J Virol. 1996;70:1890–1897. doi: 10.1128/jvi.70.3.1890-1897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perri S, Ganem D. J Virol. 1996;70:6803–6809. doi: 10.1128/jvi.70.10.6803-6809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]