Abstract
We have cloned and sequenced two defensins, Smd1 and Smd2, from anterior midgut tissue of the blood-sucking fly Stomoxys calcitrans. The DNA and N-terminal protein sequences suggest both are produced as prepropeptides. Smd1 differs from the classic defensin pattern in having an unusual six-amino acid-long N-terminal sequence. Both Smd1 and Smd2 have lower pI points and charge than insect defensins derived from fat body/hemocytes. Northern analysis shows both of these defensin molecules are tissue specific; both are produced by the anterior midgut tissue and, unlike the other insect defensins reported to date, neither appears to be expressed in fat body or hemocytes. Northern analysis also shows that mRNAs for both defensins are constitutively produced in the anterior midgut tissues and that these transcripts are up-regulated in response to sterile as well as a lipopolysaccharide-containing blood meal. However, anti-Gram-negative biological activity in the midgut is substantially enhanced by lipopolysaccharide. These findings suggest that the insect midgut has its own tissue-specific immune mechanisms and that this invertebrate epithelium is, like several vertebrate epithelia, protected by specific antibacterial peptides.
The application of modern techniques of protein biochemistry and molecular biology has led to a rapid increase in our understanding of the insect immune system (reviewed in refs. 1–4). Most work has concentrated on defense mechanisms in the hemolymph of “model” insects, and consequently our understanding of tissue-specific immune systems and immune systems in insect pest species is poor.
As with all other Metazoa the insect intestine is particularly vulnerable to attack from pathogens, parasites, and a range of opportunistic organisms ingested with the food. In particular, the midgut of blood-sucking insects is a crucial interface between potential vectors and the parasites they ingest. Given the undoubted importance to the insect of an effective midgut immune system it is surprising that so little attention has been paid to it. We do know that lectins play an antiparasitic role in the midgut of some insects (5), that peritrophic matrix has a defensive function (6), that the anti-Gram-positive enzyme lysozyme may be present, and that Plasmodium ookinetes may, on occasion, be lysed by undefined mechanisms in the midgut (7). It seems probable that other undiscovered immunological mechanisms will be involved in gut protection.
It is now widely believed that peptide antibiotic molecules play a major role in regulating natural and nonnatural flora at epithelial surfaces (8–10), and it is believed this is the role of cecropin in insect cuticular epithelia (11), α-defensins (cryptdins) in Paneth cells of the small intestine (12), and β-defensins in tracheal epithelia (13). No systematic study of peptides in the insect midgut has been undertaken although expression of cecropin has been reported in the anterior end of the larval hindgut and other larval tissues of Drosophila undergoing histolysis (14) as has slight expression of cecropin in the midgut of Bombyx mori (15). However, in both cases these molecules were expressed to a much greater degree in other tissues of the body. In this study we have investigated antibacterial activity in the midgut of adults of the stable fly Stomoxys calcitrans (a facultatively hematophagous dipteran insect), in an attempt to find antibacterial peptides that are primarily or exclusively expressed in the insect midgut epithelium in response to midgut-specific stimuli.
MATERIALS AND METHODS
Insects.
S. calcitrans were cultured in the laboratory as described previously (16). Both sexes are blood feeders, and all flies were held under laboratory culture conditions throughout the experiments. Unfed adults were collected within 24 hr of eclosion. Flies were used unfed or were fed from cotton wool-soaked swabs on either sterile, heparinized pig or rabbit blood or heparinized pig blood containing the quantities of lipopolysaccharide (LPS) (Escherichia coli serotype 026:B6; Sigma) or E. coli D31 described in the text.
Antibacterial Assay.
Anti-Gram-negative activity was determined using two separate inhibition zone assays. Nine-centimeter plates were made with 7 ml of 0.7% agarose in Luria–Bertani media containing 2 μl of a suspension of E. coli K12 RM148 grown to OD600 0.4–0.6. Plates were made with or without the inclusion of 5 mg/ml of 0.22 μM filtered lysozyme (chicken egg white, 50,000 units/mg, Sigma); E. coli K12 RM148 grows successfully in the presence of 600 mg/ml lysozyme, the maximum concentration achievable (results not shown). Addition of lysozyme increases the sensitivity of the assay (17) and permits the detection of molecules that are capable of permeabilizing the outer membrane of Gram-negative bacteria but that cannot kill them (18). One-millimeter diameter holes were punched in the set agarose and filled with 2 μl of test solution. Plates were incubated overnight at 37°C, and the diameter of inhibition zones measured. The assay was calibrated by measuring the diameter of the inhibition zone created by serial dilutions of cecropin B (Sigma) from 250 μM to 1 μM in the same assays. The calibration curve was produced by linear regression analysis of the area of the inhibition zone minus the area of the hole against log cecropin B molarity. Activity of test solutions are expressed in μM of cecropin B equivalent activity.
Preliminary Analysis of Antibacterial Activity in the Intestine.
Midguts, sections of midguts, crop, and salivary glands were dissected from unfed flies or at various times after feeding flies on heparinized pig blood containing the concentrations of LPS described in the text. Tissues were dissected in 154 mM NaCl and held at 4°C. Samples from 20 flies were homogenized in 20 μl of 0.9% NaCl, heated to 100°C for 2 min, and centrifuged at 5,000 × g, and 2 μl of the supernatant was assayed for anti-Gram-negative activity.
Purification of Antibacterial Proteins and N-Terminal Sequencing.
Flies were fed 5,000 units/ml of LPS in heparinized pig blood. Two thousand anterior midguts were dissected from those flies that had completely digested the blood meal at 24–36 hr after feeding. Tissue was dissected into 154 mM NaCl at 4°C and homogenized in a total of 1.6 ml of 200 mM sodium acetate, pH 4.5. Homogenates were heated at 100°C for 5 min and centrifuged at 9,000 × g for 15 min. The supernatant was progressively ultrafiltered through CentriCon 100, 50, and 10 filters (Amicon), separating homogenate into four groups of < 10 kDa, 10–50 kDa, 50–100 kDa, and > 100 kDa. The ultrafiltrate was centrifuged repeatedly at 4°C for 60 ≈ 180 min with 154 mM NaCl washing until retentates were minimized. After concentration by vac-centrifugation, each filtrate was subjected to reversed-phase HPLC on a C18 column (Vydac 218TP54, 4.6 × 250 mm); mobile phase A: 0.1% trifluoroacetic acid (TFA) in H2O; B: 0.1% TFA in 100% acetonitrile with a linear gradient of B from 0 to 60% over 50 min at a flow rate of 1 ml/min at ambient temperature. Peak detection was by UV at 215 nm. All peaks were collected individually and dried by vac-centrifugation. Each was resolubilized in 154 mM NaCl and tested for antibacterial activity. The purity of peptides in active peaks and the estimated molecular mass was determined by silver staining Tris-tricine-buffered SDS/PAGE gels. Nonpure, active peaks were subjected to reversed-phase HPLC on a C4 column (Vydac 214TP54, 4.6 × 250 mm) under the same conditions as those for the C18 column, except the mobile phase gradient of B was from 0 to 35% in 30 min. The activity, purity, and estimated molecular mass of purified peptides was determined as above. Occasionally, active peaks were still not pure after this second HPLC step in which case they were subjected to further purification by HPLC on a gel filtration column (BioSep SEC-2000s, 7.8 × 300 mm, Phenomenex, United Kingdom); mobile phase 20 mM phosphate buffer, pH 7.2, in 10% methanol; flow rate 0.8 ml/min at ambient temperature. The activity, purity, and estimated molecular mass of purified peptides was determined as above. Sub-samples of protein/peptides that had an unique band on SDS/PAGE gels were electrotransferred to poly(vinylidene difluoride) membrane (Immobilon, Millipore) in glycine-free buffer, pH 8.3, containing 25 mM Tris⋅HCl, and 25% methanol, or 35% methanol for peptides that were smaller than 10 kDa. Membrane was Coomassie-blue stained, and the band was cut and commercially sequenced by the Edman degradation method using an automatic protein sequencer (Applied Biosystems 473A) at Alta Bioscience (University of Birmingham, United Kingdom). For the peptides reported in this paper only one amino acid was observed per sequencing cycle. Chemicals used were HPLC grade (Supelco) for purification work and electrophoresis grade (Bio-Rad) for SDS/PAGE.
PCR Procedures.
The following degenerate oligo sense primers were synthesized commercially (GIBCO/BRL). They were designed from the N-terminal sequence information gained from the Edman degradation analysis (see Results): for Smd1, 5′-AAACCTATGGGYATHAC-3′ and for Smd2, 5′-GCTACTTGCGAYYTNYT-3′. In addition, the pBluescript M13 forward primer was used in the analysis. Fifty nanograms of template cDNA was amplified in a 100-μl reaction mixture containing, as final concentrations, 10 mM Tris⋅HCl at pH 8.3, 50 mM KCl, 1% Triton X-100, 3.5 mM MgCl2, 0.25 mM of each of the four deoxynucleotide triphosphates, 2 units of Taq DNA polymerase (GIBCO), and 1 μM each of the defensin-specific primer and M13 forward primer. A Techne FPHC3CD cyclogene thermal cycler was used as follows: 30 cycles of 94°C, 1 min; 55°C, 1 min; 72°C, 2 min followed by a single hold at 55°C for 5 min. PCR products were visualized after electrophoresis through a 1% agarose gel containing 0.5 μg/ml ethidium bromide.
To generate probes for library screening and Northern analysis, PCR products were purified and digested with XhoI to remove vector. Probes for Northern analysis also were made from purified plasmids containing full-length inserts (Qiagen plasmid kit) by restriction with EcoRI and XhoI. Probes were 32P-labeled using random primers (Appligene Nona Primer kit).
cDNA Library Construction, Screening, and Sequencing of Selected Clones.
To avoid problems arising from the presence of mRNA in blood, female S. calcitrans were fed an artificial meal consisting of 15 mg/ml porcine γ-globulin, 8 mg/ml hemoglobin, 177 mg/ml albumin, 9.0 mg/ml NaCl, 1.7 mg/ml NaHCO3, and 0.55 mg/ml ATP. This formulation induced comparable levels of trypsin activity in the midgut to that induced by whole porcine blood (data not shown). Eight hundred midguts were dissected 6–12 hr postblood meal in 154 mM NaCl and immediately frozen in liquid N2.
Poly(A)+ RNA was isolated (Dynal mRNA Direct kit). cDNA was synthesized, directionally cloned, and packaged (ZAP-cDNA Synthesis Kit, Stratagene). The library, which was estimated to contain 2.7 × 106 independent clones, was plated using E. coli XL1-Blue and screened with a 32P-dCTP-labeled probe derived from PCR products purified by electroelution. pBluescript phagemids were excised in vivo from the lambda vector using ExAssist helper phage and plated using the E. coli SOLR. Dideoxy DNA sequencing was carried out using the Pharmacia T7 polymerase kit.
Northern Analysis.
Adult flies of mixed sex, previously unfed and 12–36 hr since eclosion, were fed either sterile heparinized pig blood or heparinized pig blood plus 3,000 units/ml of LPS. Other flies were injected with 50 ng LPS (2.5 units) in 1 μl of sterile 154 mM NaCl through the pteropleural region of the thorax 24 hr after feeding on sterile rabbit blood. Flies were killed by freezing in liquid N2 at the times indicated in the text. Tissues were dissected 24–30 hr postblood meal in 154 mM NaCl and immediately frozen in liquid N2. The tissues were separated into anterior midgut, posterior midgut, abdominal fat body, and the remainder of the body. Total RNA was extracted from flies or tissues by the acid guanidinium thiocyanate/phenol/chloroform method (19). Thirty or 50 micrograms of whole-body RNA or 10 μg of RNA from specific tissues were used for Northern analysis. RNA was denatured and electrophoresed through a 1.3% agarose/formaldehyde gel and transferred to Biotrans nylon membrane (ICN). Membranes were prehybridized at 42°C for 5 hr in 50% (vol/vol) formamide/5× Denhardt’s solution/5× standard saline citrate (SSC)/0.1% SDS/100 μg/ml salmon sperm DNA. Hybridization was for 36 hr at 42°C in the same solution with the addition of 100 μg/ml poly(A) and 32P-labeled Smd1 or Smd2 defensin or Aedes aegypti ribosomal probes (gift of J. Crampton, University of Liverpool, U.K.). After hybridization the membrane was sequentially washed for 2 × 10-min periods in 2× SSC/0.1% SDS; 2 × 10 min in 1× SSC/0.1% SDS both at room temperature followed by 2 × 10 min in 0.1× SSC/0.1% SDS at 42°C and 2 × 10-min washes in 0.1× SSC/0.1% SDS at 65°C. Membranes were exposed to Kodak X-Omat film for 24–72 hr at −50°C. Densitometry was performed on an LKB 2202 laser densitometer.
RESULTS
Inducible Antibacterial Activity in the Midgut.
Anti-Gram-negative activity is apparent in crude homogenates of fly midguts and peaks 24–36 hr after a blood meal (results not shown). To determine which of the functionally distinct zones of the midgut possesses anti-Gram-negative activity, crude homogenates of the different zones were assayed separately. Activity was found throughout the entire anterior midgut, whereas neither the posterior midgut nor the salivary glands showed activity (Fig. 1). The activity of the anterior midgut increases with doses of LPS exceeding approximately 2,000 units per ml of LPS (Fig. 2), suggesting this may represent a response threshold.
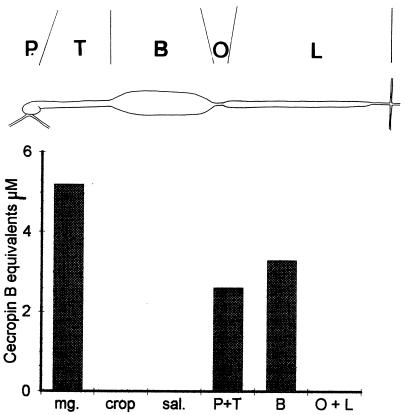
Figure 1.
Segregation of anti-Gram-negative activity in the midgut. The anterior midgut of adult S. calcitrans is divided into proventriculus (P), thoracic midgut (T), and reservoir (B). The posterior midgut is formed of the opaque zone (O) and the lipoid zone (L). The anti-Gram-negative activity of these various regions of the midgut of mixed sex flies 24 hr after feeding on a 50/50 mixture of pig blood and E. coli D31 grown to 0.55 at OD600 assayed in the presence of lysozyme is presented. Two gut equivalents per assay point. mg, total midgut; crop, crop; sal, salivary glands.
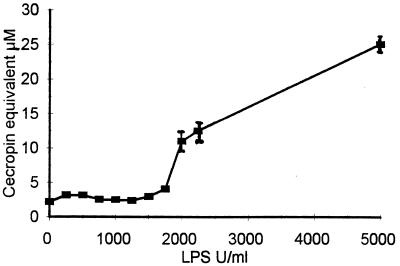
Figure 2.
Means ± SE of anti-Gram-negative activity, assayed in the presence of lysozyme, in the supernatants of homogenized anterior midguts of mixed sex flies (previous analysis of variance showed no significant difference between the sexes, P = 0.895, n = 24) 36 hr after feeding on heparinized pig blood containing the indicated doses of LPS. Two gut equivalents per assay point.
Purification and N-Terminal Sequencing.
Screening of HPLC-purified proteins revealed a number displaying anti-Gram-negative activity, and the further study of two of these is reported here. The first, termed Smd1 (Stomoxys midgut defensin 1), was present in the final HPLC fraction at a concentration of 8 μM, and when assayed at this concentration gave no activity in the assay without lysozyme but in the assay with lysozyme gave activity equivalent to 2.5 μM cecropin B. The second, termed Smd2 (Stomoxys midgut defensin 2), was present in the final HPLC fraction at a concentration of 24 μM, and when assayed at this concentration showed activity in the assay without lysozyme equivalent to 13.4 μM cecropin B and in the assay with lysozyme activity equivalent to 19 μM cecropin B. Edman degradation analysis gave an N-terminal sequence of AAKPMGITXDLLXL for Smd1 and ATXDLLXMXN for Smd2.
PCR Analysis.
A PCR primer was designed for positions 3–8 (number refers to Edman sequence given above) in Smd1. Reference to consensus sequences for insect defensins (3) suggested position 3 in Smd2 could be C. Making this assumption, we designed a PCR primer for positions 1–6 in Smd2. PCR reactions of Smd1 using the cDNA library as template gave reaction products of approximately 490 and 290 bp. Preliminary sequencing of the 490-bp band suggested the product coded for a 40S ribosomal S14 protein, and this product was abandoned. The 290-bp product was used as a probe for library screening and Northern analysis. PCR reactions of Smd2 gave a reaction product of 280 bp, which was used as a probe for library screening and Northern analysis.
Cloning and Sequencing.
Clones were selected and sequenced; the nucleotide and amino acid sequence of the full-length clones of Smd1 and Smd2 are given in Fig. 3. Smd1 has an ORF of 189 nucleotides coding for a peptide of 79 residues with a calculated molecular mass of 8,159 Da. Smd2 has an ORF of 291 nucleotides coding for a peptide of 97 residues with a calculated molecular mass of 10,614 Da.
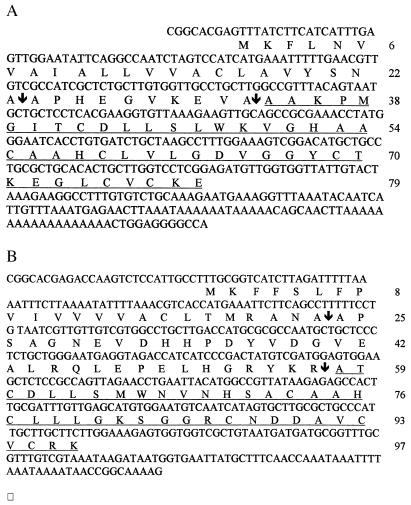
Figure 3.
The nucleotide sequence of the two cDNA clones and the deduced amino acid sequences of the two peptides. Amino acid residues are numbered from the first methionine residue. The mature peptide, as determined from the Edman degradation studies, is underlined, and the arrows indicate the putative cleavage sites of the signal sequence and pro sequence. (A) Smd1. (B) Smd2.
Northern Analysis.
After feeding on LPS-containing blood was completed, Northern analysis shows Smd1 and Smd2 are located only in the anterior midgut (Fig. 4A). The injection of LPS into the hemocoel of another dipteran, Phormia terranovae, induces transcription of defensin molecules in fat body and hemocytes (20). We injected LPS into the hemocoel of S. calcitrans and assume this also will induce transcription of fat body and hemocyte defensins. However, under these conditions Smd1 and Smd2 are still found only in the anterior midgut with no labeling of the fat or remainder of the body lanes (Fig. 4B). These results suggest mRNA for Smd1 and Smd2 is produced exclusively in anterior midgut tissues of adult S. calcitrans. Washing the Northerns of specific tissues less stringently to 0.1× SSC/0.1% SDS at 42°C gave no further banding when probed with Smd1, but when probed with Smd2 a faint band of hybridization appeared in the remainder of the body lane (results not shown). This band was completely lost when the membrane was washed to 0.1× SSC/0.1% SDS at 65°C. This suggests either very low levels of transcription of Smd2 or transcription of a closely related, but not identical, mRNA somewhere else in the body but not in the fat body. Northern analysis shows constitutive expression of Smd1 and Smd2, which is up-regulated by blood feeding (Fig. 4C). Fig. 4 C and D compares the effects of a sterile blood meal and a blood meal containing LPS at a concentration of LPS that is known to increase anti-Gram-negative activity (Fig. 2), on the levels of Smd1 and Smd2. Densitometry of these Northerns showed that LPS caused peak levels of Smd1 to be depressed by about 30%, whereas it enhanced peak levels of Smd2 by about 30%. Presence of LPS also caused a more prolonged increase in Smd2 levels with over 80% more mRNA present at 48 hr postblood meal.
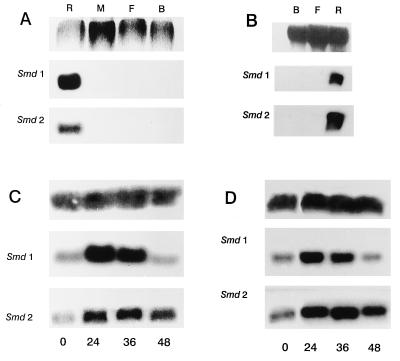
Figure 4.
Northern analysis. Each of the membranes (A-D) were probed once with Smd1, stripped, and reprobed with Smd2. (A) Specific tissues from flies fed on pig blood containing 3,000 units/ml LPS. (Upper) Ethidium bromide staining of RNA. (B) Specific tissues from flies injected with LPS. (Upper) Methylene blue staining of RNA. (C) Whole fly extracts at the indicated hours after feeding on sterile, heparinized pig blood. (Upper) Aedes aegypti ribosomal probe. (D) Whole fly extracts at the indicated hours after feeding on heparinized pig blood plus LPS at 3,000 units/ml. (Upper) A. aegypti ribosomal probe. R, anterior midgut; M, posterior midgut; F, fat body; B, remainder of the carcass.
DISCUSSION
Anti-Gram-negative activity is restricted to the anterior midgut, which is the region where peritrophic matrix is produced (21) and where blood is stored and dehydrated (22) before the addition of digestive enzymes from more posterior regions of the midgut where digestion and absorption occur (21, 23–26). Undigested blood, which may be stored for up to 48 hr in this region, is a potentially rich medium for bacterial growth. The antibacterial activity that we have demonstrated is centered in this region of gut may help prevent bacterial growth in this valuable resource.
It is tentatively suggested that both Smd1 and Smd2 are preproproteins. The putative leader sequence comprises the first 23 amino acids of Smd1 terminating in Ala, which is typical of this type of peptide (20, 27–29). If the Edman degradation analysis was indeed of the mature peptide this suggests the pro-sequence is 10 residues long and ends in another Ala residue. Given this assumption the mature peptide is 46 amino acids in length with a calculated molecular mass of 4,736 Da. It is tentatively suggested that the leader sequence of Smd2 comprises the first 23 amino acids and that cleavage occurs within the same amino acid sequence, Asn-Ala-Ala-Pro, as Smd1. If this is correct we suggest the pro-sequence is 34 residues long and ends in the classic proteolytic cleavage site Lys-Arg used for other defensins (20, 29), but not for Smd1. This gives a mature peptide 40 amino acids in length with a calculated molecular mass of 4,237 Da.
Smd1 and Smd2 are both defensins, although Smd1 is more unusual relative to previously characterized members of the family (Fig. 5). The six cysteine residues in Smd1 and Smd2 are in the positions that are invariant in all mature insect defensins studied. The putative leader sequences of Smd1 and Smd2 are typical in containing a large number of hydrophobic amino acids (Fig. 3). With the exception of the putative cleavage site, little conservation of amino acid identity is in the leader sequence of either Smd1 or Smd2 compared with other insect defensins (Fig. 5). Within the pro-sequence of Smd2 the 11 amino acids N-terminal to the putative activation site are identical to those of P. terranovae isoform A (30) and S. peregrina sapecin isoform A (31) with further identical sequences N-terminal of this region. In contrast, the putative pro-sequence of Smd1 is unusually short and shows no similarity to the pro-sequence of any other defensin studied. In addition, whereas the putative cleavage site of the pro-sequence from the mature peptide in Smd2 is typical of other defensins that of Smd1 is atypical. The significance of these differences remains to be determined, but the variation in the putative cleavage site of the pro-sequence from the mature peptide site in particular is consistent with a novel processing sequence for Smd1. The sequences of mature Smd1 and Smd2 peptides show a high degree of identity to other insect defensins (Fig. 5) with the exception being six extra amino acids at the N terminus of Smd1. The significance of this atypical N-terminal sequence remains to be determined, but it is it worth noting that the vertebrate Paneth cell cryptdins also have amino termini that are 4–6 residues longer than the average amino terminus of the leukocyte-derived α-defensins (32). With the possibility the peptide may function in the midgut lumen it also is interesting to note that Apis mellifera royalisin (33) and the defensin-related charybdotoxin from Leiurus quinquestriatus (34), both of which operate outside the body of the arthropod, also have respectively atypical C-terminal and N-terminal extensions from the standard defensin-type pattern. With the exception of the first six amino acids in mature Smd1 none of the amino acids distinguishing S. calcitrans defensins from the other insect defensins studied are of obvious significance in determining higher-order structural features. However, differences exist in pI and charge of the molecules at pH 7, which separate these midgut defensins from fat body/hemocyte-derived defensins. Mature Smd1 and Smd2 have a calculated pI and charge of 7.05, +0.06 and 7.66, +1.06, respectively. This compares with means (ranges) of 8.29 (8.02–8.53), +2.96 (+1.89–+3.89) for the mature defensins presented in Fig. 5. The physiological significance of these differences remains to be determined.
Figure 5.
Smd1 and Smd2 are compared with defensins from the orders Diptera, Hymenoptera, Coleoptera, and Odonata using single-letter notation for amino acids. Where sequences have been reported the full putative preprodefensin is presented for comparison, otherwise only the mature defensins are compared. The putative pre-, pro-, and mature peptides are presented separately. Ac, Aeschna cyanea (38); Ae, Aedes aegypti (39); Am, Apis mellifera (33); An, Anopheles gambiae (29); Dm, Drosophila melanogaster (40); Pt, Phormia terranovae isoform A (30); Sp, Sarcophaga peregrina sapecin A (31); Tm, Tenebrio molitor (41); Za, Zophobas atratus isoform A (42). Residues identical to Smd2 are represented by dots in the sequence column. Residues that are identical to Smd1 are shaded. Gaps introduced by the alignment program are represented by dashes. The six conserved cysteine residues in the mature peptide are indicated by ∗. Sequences were aligned using clustal v.
The evidence suggests that defensins Smd1 and Smd2 are specific to the anterior midgut tissues of S. calcitrans. First, these defensins have been cloned from an anterior midgut-specific cDNA library. It is, of course, possible, but in our opinion unlikely, that tissues other than midgut were contaminants in the production of the cDNA library. The midgut-specific nature of these defensins is supported by Northern analysis. The injection of LPS is very likely to promote the expression of defensins in fat body and blood cells (20), but these are not recognized by our defensin probes, which only bind in the anterior midgut lane (Fig. 4). The finding of a weak band of staining for Smd2 in the remainder of the body lane when less stringent washing conditions were used suggests either low levels of Smd2 are produced elsewhere or that a related, but not identical, defensin is to be found in other tissues of the body. The evidence suggests these other tissues do not include abdominal fat body or posterior midgut.
We report the identification of the source of insect defensins from tissues other than fat body and hemocytes. The finding is consistent with the view that insect midgut may possess its own particular immune molecules. It is worth noting that in vertebrates the α-defensin genes expressed in Paneth cells and cells of myeloid lineage are also distinct from each other. Enteric defensin RNA does not crosshybridize with myeloid cell RNA (35) and vice versa (36).
Anterior midgut tissue is a mixture of midgut epithelium and small numbers of putative endocrine cells surrounded by muscles and trachea (M.J.L., unpublished work). Which cell type is producing defensin mRNA is not known. The Edman sequencing data taken together with the sequencing of the clones is evidence that the defensins are produced as preproproteins, which is consistent with a secretory fate. The eventual location of the mature proteins has not been established, although it is tempting to speculate that these insect defensins, like for example the cryptdins of Paneth cells (37), may be secreted into the gut lumen.
In fat body/hemocyte tissues defensin mRNA is usually undetectable in unchallenged insects (20) or where it is detectable at low levels this is assumed to reflect background injuries or infections (29). In contrast, the Northern analysis of midgut defensins reported here (Fig. 4) shows that mRNA is present in unstimulated insects. It is, of course, possible that the midgut per se is not sterile. However, unfed insects had received no food as adults and so any stimulant is most likely to have arisen from larval feeding, more than 6 days previously. The evidence suggests that defensin mRNA is continually present in these tissues independent of challenge. Such transcripts would permit a rapid response to challenge, a potentially important factor in these tissues that form part of the front line of the insect’s defense. In this regard it is interesting to note that in the vertebrate gut cryptdins from Paneth cells are secreted into the lumen on a constitutive basis (12).
A certain level of Smd1 and Smd2 transcripts are found in the midgut constitutively. However, both a sterile blood meal and a meal containing LPS increase the levels of Smd1 and Smd2 (Fig. 4 C and D). Addition of LPS to the blood meal at the same level does lead to an increase in anti-Gram-negative activity in the midgut (Fig. 2); the anterior midgut shows Smd1 and Smd2 transcripts both after feeding on LPS-containing blood (Fig. 4A) and when the fly is injected with LPS (Fig. 4B). Additional experiments are required to determine whether LPS may induce transcription as well as activate posttranscriptionally RNAs for defensin and other antibacterial peptides.
In summary our evidence suggests that the defensins Smd1 and Smd2 are specific to anterior midgut tissue in which they are constitutively expressed. These defensins are only two of a number of antibacterial molecules that are present in the anterior midgut of adult S. calcitrans (data not shown).
Note Added in Proof
Anterior midgut specificity of defensin expression and induction of multiple immune markers in the midgut by malaria have been documented in the mosquito Anopheles gambiae by Dimopoulos et al. (43).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: LPS, lipopolysaccharide.
Data deposition: The nucleotide sequences of Smd1 and Smd2 have been deposited with the GenBank database (accession nos. AF013146 and AF013147, respectively).
A commentary on this article begins on page 11152.
References
- 1.Boman H G, Hultmark D. Annu Rev Microbiol. 1987;41:103–126. doi: 10.1146/annurev.mi.41.100187.000535. [DOI] [PubMed] [Google Scholar]
- 2.Boman H G. Cell. 1991;65:205–207. doi: 10.1016/0092-8674(91)90154-q. [DOI] [PubMed] [Google Scholar]
- 3.Cociancich S, Bulet P, Hetru C, Hoffmann J A. Parasitol Today. 1994;10:132–139. doi: 10.1016/0169-4758(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 4.Ham P J, Albuquerque C, Smithies B, Chalk R, Klager S, Hagen H. In: Antimicrobial Peptides, Ciba Foundation Symposia 186. Marsh J, Goode J A, Boman H, editors; Marsh J, Goode J A, Boman H, editors. Chichester, U.K.: Wiley; 1994. pp. 140–151. [DOI] [PubMed] [Google Scholar]
- 5.Maudlin I. Adv Dis Vect Res. 1991;7:117–149. [Google Scholar]
- 6.Lehane M J. Annu Rev Entomol. 1997;42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- 7.Vernick K D, Fujioka H, Seeley D C, Tandler B, Aikawa M, Miller L H. Exp Parasitol. 1995;80:583–595. doi: 10.1006/expr.1995.1074. [DOI] [PubMed] [Google Scholar]
- 8.Diamond G, Jones D E, Bevins C L. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bevins C L. In: Antimicrobial Peptides, Ciba Foundation Symposium 186. Marsh J, Goode J A, Boman H, editors; Marsh J, Goode J A, Boman H, editors. Chichester, U.K.: Wiley; 1994. pp. 250–269. [DOI] [PubMed] [Google Scholar]
- 10.Boman H G. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 11.Brey P T, Lee W J, Yamakawa M, Koizumi Y, Perrot S, Francois M, Ashida M. Proc Natl Acad Sci USA. 1993;90:6275–6279. doi: 10.1073/pnas.90.13.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selsted M E, Miller S I, Henschen A H, Ouellette A J. J Cell Biol. 1992;118:929–936. doi: 10.1083/jcb.118.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamond G, Jones D E, Bevins C L. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tryselius Y, Samakovlis C, Kimbrell D A, Hultmark D. Eur J Biochem. 1992;204:395–399. doi: 10.1111/j.1432-1033.1992.tb16648.x. [DOI] [PubMed] [Google Scholar]
- 15.Yamano Y, Matsumoto M, Inoue K, Kawabata T, Morishima I. Biosci Biotechnol Biochem. 1994;58:1476–1478. doi: 10.1271/bbb.58.1476. [DOI] [PubMed] [Google Scholar]
- 16.Blakemore D, Lehane M J, Williams S. Insect Biochem Mol Biol. 1993;23:331–335. [Google Scholar]
- 17.Chalk R, Townson H, Natori S, Desmond H, Ham P J. Insect Biochem Mol Biol. 1994;24:403–410. doi: 10.1016/0965-1748(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 18.Vaara M. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Dimarcq J-L, Zachary D, Hoffmann J A, Hoffmann D, Reichhart J-M. EMBO J. 1990;9:2507–2515. doi: 10.1002/j.1460-2075.1990.tb07430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehane M J. Cell Tissue Res. 1976;170:275–287. doi: 10.1007/BF00224304. [DOI] [PubMed] [Google Scholar]
- 22.Macvicker J A K, Billingsley P F, Djamgoz M B A, Harrow I D. Insect Biochem Mol Biol. 1994;24:151–159. [Google Scholar]
- 23.Lehane M J. J Insect Physiol. 1977;23:945–954. doi: 10.1016/0022-1910(77)90121-4. [DOI] [PubMed] [Google Scholar]
- 24.Lehane M J. Tissue Cell. 1989;21:101–111. doi: 10.1016/0040-8166(89)90025-6. [DOI] [PubMed] [Google Scholar]
- 25.Jordao B P, Lehane M J, Terra W R, Ribeiro A F, Ferreira C. Insect Biochem Mol Biol. 1996;26:445–453. doi: 10.1016/0965-1748(95)00112-3. [DOI] [PubMed] [Google Scholar]
- 26.Moffatt M R, Blakemore D, Lehane M J. Comp Biochem Physiol B. 1995;110:291–300. [Google Scholar]
- 27.Lidholm D A, Gudmundsson G H, Xanthopoulos K G, Boman H G. Febs Lett. 1987;226:8–12. [Google Scholar]
- 28.Matsumoto N, Okada M, Takahashi H, Ming Q X, Nakajima Y, Nakanishi Y, Komano H, Natori S. Biochem J. 1986;239:717–722. doi: 10.1042/bj2390717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richman A M, Bulet P, Hetru C, Barillas-Mury C, Hoffmann J A, Kafatos F C. Insect Mol Biol. 1996;5:203–210. doi: 10.1111/j.1365-2583.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 30.Lambert J, Keppi E, Dimarcq J-L, Wicker C, Reichhart J-M, Dunbar B, Lepage P, van Dorsselaer J A, Fothergill J, Hoffmann D. Proc Natl Acad Sci USA. 1989;86:262–266. doi: 10.1073/pnas.86.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama K, Natori S. J Biol Chem. 1988;263:17117–17121. [PubMed] [Google Scholar]
- 32.Ouellette A J, Selsted M E. FASEB J. 1996;10:1280–1289. doi: 10.1096/fasebj.10.11.8836041. [DOI] [PubMed] [Google Scholar]
- 33.Casteels-Josson K, Zhang W, Capaci T, Casteels P, Tempst P. J Biol Chem. 1994;269:28569–28575. [PubMed] [Google Scholar]
- 34.Miller C, Moczydlowski E, Latorre R, Phillips M. Nature (London) 1985;313:316–318. doi: 10.1038/313316a0. [DOI] [PubMed] [Google Scholar]
- 35.Jones D E, Bevins C L. FEBS Lett. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 36.Linzmeier R, Michaelson D, Liu L, Ganz T. FEBS Lett. 1993;321:267–273. doi: 10.1016/0014-5793(93)80122-b. [DOI] [PubMed] [Google Scholar]
- 37.Selsted M E, Ouellette A J. Trends Cell Biol. 1995;5:114–119. doi: 10.1016/s0962-8924(00)88961-8. [DOI] [PubMed] [Google Scholar]
- 38.Bulet P, Cociancich S, Reuland M, Sauber F, Bischoff R, Hegy G, Van D A, Hetru C, Hoffmann J A. Eur J Biochem. 1992;209:977–984. doi: 10.1111/j.1432-1033.1992.tb17371.x. [DOI] [PubMed] [Google Scholar]
- 39.Lowenberger C, Bulet P, Charlet M, Hetru C, Hodgeman B, Christensen B M, Hoffmann J A. Insect Biochem Mol Biol. 1995;25:867–873. doi: 10.1016/0965-1748(95)00043-u. [DOI] [PubMed] [Google Scholar]
- 40.Dimarcq J L, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart J M, Hoffmann J A. Eur J Biochem. 1994;221:201–209. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- 41.Moon H J, Lee S Y, Shoichiro K, Natori S, Lee B L. J Biochem. 1994;116:53–58. doi: 10.1093/oxfordjournals.jbchem.a124502. [DOI] [PubMed] [Google Scholar]
- 42.Bulet P, Cociancich S, Dimarcq J L, Lambert J, Reichhart J M, Hoffmann D, Hetru C, Hoffmann J A. J Biol Chem. 1991;266:24520–24525. [PubMed] [Google Scholar]
- 43.Dimopoulos G, Richman A, Müller H-M, Kafatos F C. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]