Abstract
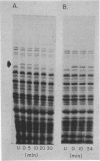
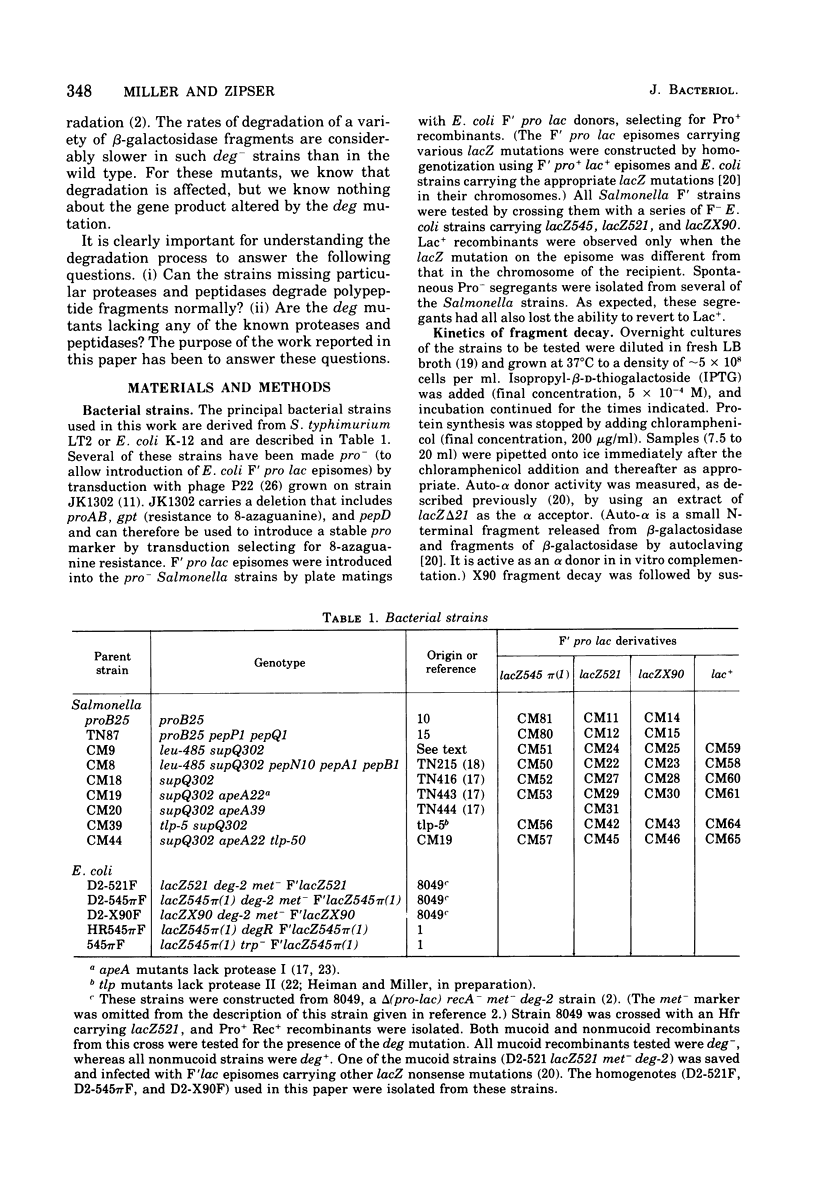
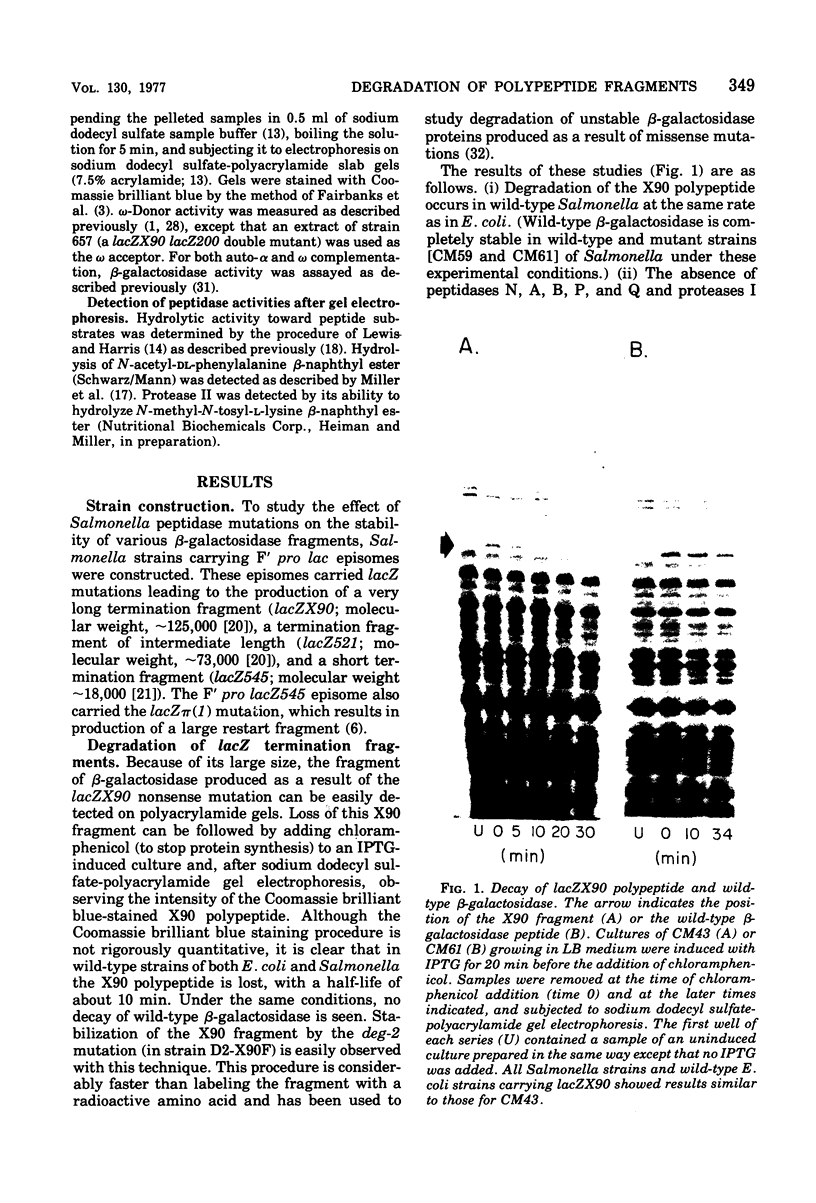
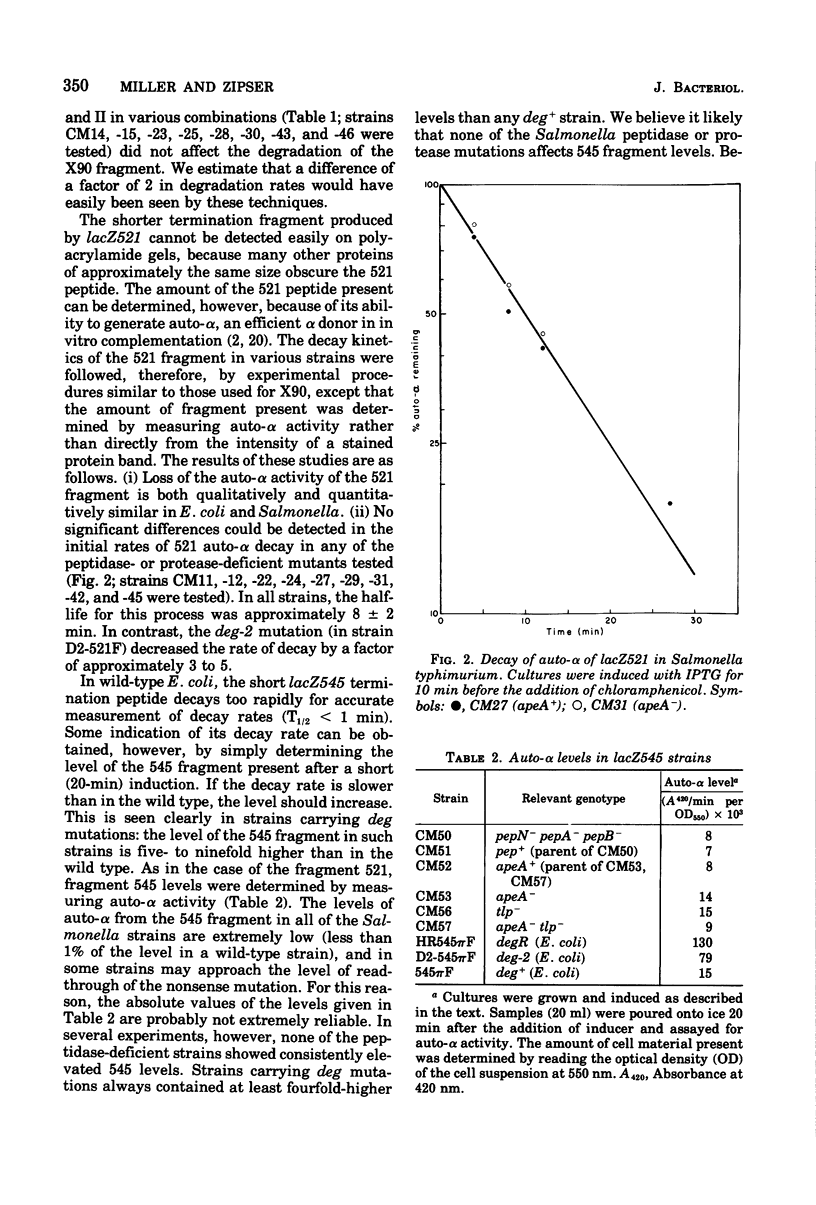
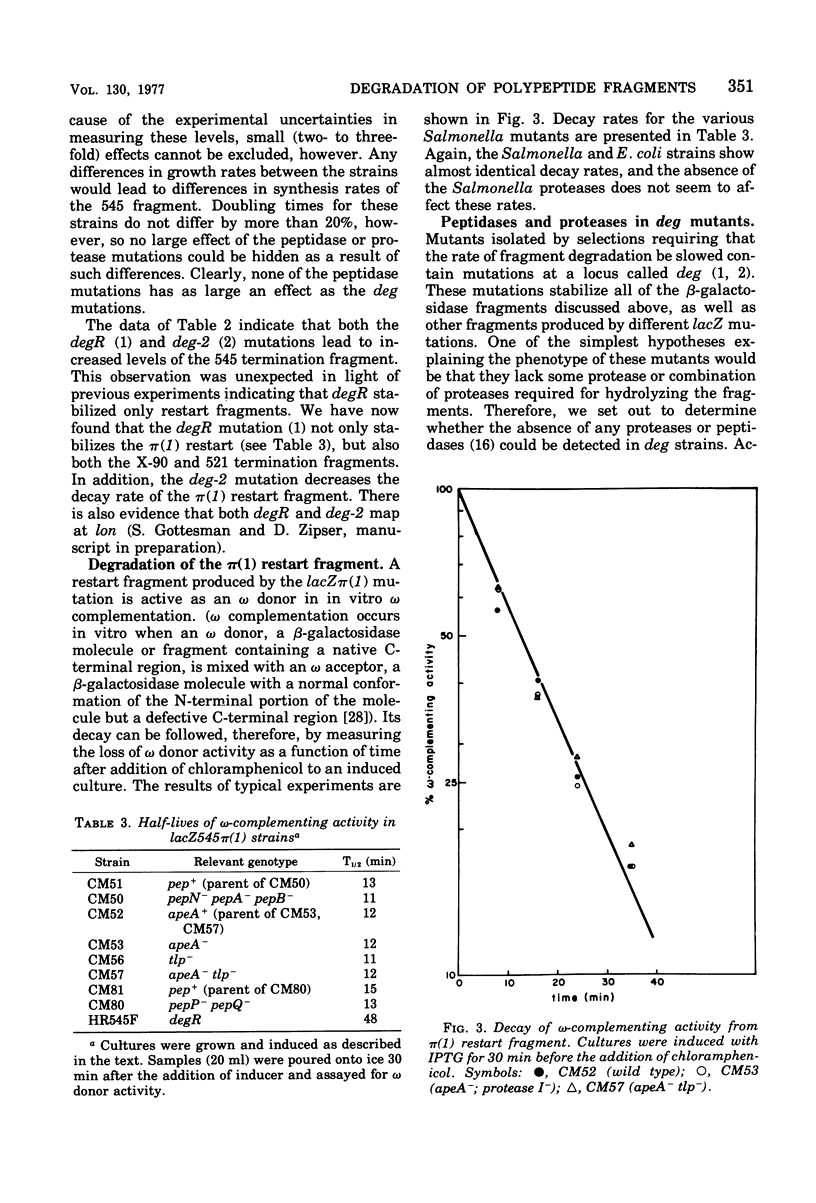
The degradation rates of several mutationally generated fragments of Escherichia coli beta-galactosidase were determined in wild-type strains of Salmonella typhimurium and in mutant Salmonella strains lacking several proteases and peptidases. Three termination fragments (produced by lacZ545, lacZ521, and lacZX90) and one internal reinitiation (restart) fragment [lacZpi(1)] are degraded in wild-type Salmonella strains at the same rates observed in wild-type Escherichia coli strains. Mutations that lead to loss of peptidases N, A, B, P, and Q or to loss of protease I or II do not affect the decay rates of any of these fragments. In addition, all of these peptidases and proteases are present in E coli mutants carrying deg mutations (deg mutations are known to stabilize beta-galactosidase fragments). Apparently, none of the proteases and peptidases that are currently accessible to direct genetic analysis plays a role in the early steps of the degradation of protein fragments.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apte B. N., Rhodes H., Zipser D. Mutation blocking the specific degradation of reinitiation polypeptides in E. coli. Nature. 1975 Sep 25;257(5524):329–331. doi: 10.1038/257329a0. [DOI] [PubMed] [Google Scholar]
- Bukhari A. I., Zipser D. Mutants of Escherichia coli with a defect in the degradation of nonsense fragments. Nat New Biol. 1973 Jun 20;243(129):238–241. doi: 10.1038/newbio243238a0. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. Degradation of abnormal proteins in Escherichia coli (protein breakdown-protein structure-mistranslation-amino acid analogs-puromycin). Proc Natl Acad Sci U S A. 1972 Feb;69(2):422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L., St John A. C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. In vivo degradation of nonsense fragments in E. coli. Nature. 1970 Dec 19;228(5277):1151–1154. doi: 10.1038/2281151a0. [DOI] [PubMed] [Google Scholar]
- Grodzicker T., Zipser D. A mutation which creates a new site for the re-initiation of polypeptide synthesis in the z gene of the lac operon of Escherichia coli. J Mol Biol. 1968 Dec;38(3):305–314. doi: 10.1016/0022-2836(68)90388-4. [DOI] [PubMed] [Google Scholar]
- Hermier B., Pacaud M., Dubert J. M. Action of protease I of Escherichia coli on RNA polymerase of same origin. Eur J Biochem. 1973 Oct 5;38(2):307–310. doi: 10.1111/j.1432-1033.1973.tb03063.x. [DOI] [PubMed] [Google Scholar]
- Itikawa H., Demerec M. Salmonella typhimurium proline mutants. J Bacteriol. 1968 Mar;95(3):1189–1190. doi: 10.1128/jb.95.3.1189-1190.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper J. Evolution of a new gene substituting for the leuD gene of Salmonella typhimurium: origin and nature of supQ and newD mutations. J Bacteriol. 1974 Dec;120(3):1176–1185. doi: 10.1128/jb.120.3.1176-1185.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowit J. D., Choy W. N., Champe S. P., Goldberg A. L. Role and location of "protease I" from Escherichia coli. J Bacteriol. 1976 Dec;128(3):776–784. doi: 10.1128/jb.128.3.776-784.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazdunski C., Busuttil J., Lazdunski A. Purification and properties of a periplasmic aminoendopeptidase from Escherichia coli. Eur J Biochem. 1975 Dec 15;60(2):363–369. doi: 10.1111/j.1432-1033.1975.tb21011.x. [DOI] [PubMed] [Google Scholar]
- Lewis W. H., Harris H. Human red cell peptidases. Nature. 1967 Jul 22;215(5099):351–355. doi: 10.1038/215351a0. [DOI] [PubMed] [Google Scholar]
- Lin S., Zabin I. Beta-galactosidase. Rates of synthesis and degradation of incomplete chains. J Biol Chem. 1972 Apr 10;247(7):2205–2211. [PubMed] [Google Scholar]
- McHugh G. L., Miller C. G. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):364–371. doi: 10.1128/jb.120.1.364-371.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G., Heiman C., Yen C. Mutants of Salmonella typhimurium deficient in an endoprotease. J Bacteriol. 1976 Jul;127(1):490–497. doi: 10.1128/jb.127.1.490-497.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G., Mackinnon K. Peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):355–363. doi: 10.1128/jb.120.1.355-363.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. G. Peptidases and proteases of Escherichia coli and Salmonella typhimurium. Annu Rev Microbiol. 1975;29:485–504. doi: 10.1146/annurev.mi.29.100175.002413. [DOI] [PubMed] [Google Scholar]
- Morrison S. L., Zipser D., Goldschmidt R. Polypeptide products of nonsense mutations. II. Minor fragments produced by nonsense mutations in the z gene of the lactose operon of Escherichia coli. J Mol Biol. 1971 Sep 28;60(3):485–497. doi: 10.1016/0022-2836(71)90183-5. [DOI] [PubMed] [Google Scholar]
- Morrison S. L., Zipser D. Polypeptide products of nonsense mutations. I. Termination fragments from nonsense mutations in the Z gene of the lac operon of Escherichia coli. J Mol Biol. 1970 Jun 14;50(2):359–371. doi: 10.1016/0022-2836(70)90198-1. [DOI] [PubMed] [Google Scholar]
- Pacaud M., Richaud C. Protease II from Escherichia coli. Purification and characterization. J Biol Chem. 1975 Oct 10;250(19):7771–7779. [PubMed] [Google Scholar]
- Pacaud M., Uriel J. Isolation and some propeties of a proteolytic enzyme from Escherichia coli (protease I). Eur J Biochem. 1971 Dec 10;23(3):435–442. doi: 10.1111/j.1432-1033.1971.tb01638.x. [DOI] [PubMed] [Google Scholar]
- Pine M. J. Response of intracellular proteolysis to alteration of bacterial protein and the implications in metabolic regulation. J Bacteriol. 1967 May;93(5):1527–1533. doi: 10.1128/jb.93.5.1527-1533.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Miller J. H., Weber K. In vivo degradation of mutant lac repressor. Nature. 1970 Dec 19;228(5277):1154–1156. doi: 10.1038/2281154a0. [DOI] [PubMed] [Google Scholar]
- Ullmann A., Perrin D., Jacob F., Monod J. Identification par complémentation in vitro et purification d'un segment peptidique de la beta-galatosidase d'escherichia coli. J Mol Biol. 1965 Jul;12(3):918–923. doi: 10.1016/s0022-2836(65)80338-2. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and properties of an aminopeptidase from Escherichia coli. J Biol Chem. 1970 Sep 25;245(18):4760–4769. [PubMed] [Google Scholar]
- ZIPSER D. A STUDY OF THE UREA-PRODUCED SUBUNITS OF BETA-GALACTOSIDASE. J Mol Biol. 1963 Aug;7:113–121. doi: 10.1016/s0022-2836(63)80040-6. [DOI] [PubMed] [Google Scholar]
- Zipser D., Bhavsar P. Missense mutations in the lacZ gene that result in degradation of beta-galactosidase structural protein. J Bacteriol. 1976 Sep;127(3):1538–1542. doi: 10.1128/jb.127.3.1538-1542.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]