Abstract
The factors that regulate the perpetuation and invasiveness of rheumatoid synovitis have been the subject of considerable inquiry, and the possibility that nonimmunologic defects can contribute to the disease has not been rigorously addressed. Using a mismatch detection system, we report that synovial tissue from the joints of severe chronic rheumatoid arthritis patients contain mutant p53 transcripts, which were not found in skin samples from the same patients or in joints of patients with osteoarthritis. Mutant p53 transcripts also were identified in synoviocytes cultured from rheumatoid joints. The predicted amino acid substitutions in p53 were identical or similar to those commonly observed in a variety of tumors and might influence growth and survival of rheumatoid synoviocytes. Thus, mutations in p53 and subsequent selection of the mutant cells may occur in the joints of patients as a consequence of inflammation and contribute to the pathogenesis of the disease.
Rheumatoid arthritis (RA) is a chronic inflammatory disease, and the factors that contribute to its onset, perpetuation, and invasiveness have been extensively studied. However, the possible role of nonimmunologic factors in the progression of RA has not been rigorously addressed. Recent evidence suggests that the destructive components of chronic rheumatoid synovitis can progress independently of the inflammatory elements (1–4). One possible explanation for the transition of RA to an autonomous process is that the cells that line the joint (fibroblast-like synoviocytes) achieve a degree of independent activation that results in accelerated extracellular matrix destruction. In support of this notion, considerable data have accrued indicating that fibroblast-like synoviocytes exhibit a transformed phenotype that might account for many features of established RA (5–7). However, the mechanism of synoviocyte “transformation” remains unknown.
In recent studies of the RA synovium, a discrepancy has been found between excessive DNA strand breaks and minimal apoptotic morphology in the intimal lining cells (8, 9). Marked overexpression of the p53 tumor suppressor gene subsequently was observed in the region of DNA fragmentation (10). Because high p53 expression has been associated with point mutations and abnormal apoptosis (11–13), we hypothesized that alterations in the structure or function of p53 might contribute to the transformed phenotype of RA synoviocytes. We now present data demonstrating that somatic mutations occur in the p53 tumor suppressor gene in RA synovium and in cultured rheumatoid fibroblast-like synoviocytes.
METHODS
Patient Selection and Tissue Preparation.
Synovial tissue was collected at the time of joint replacement from patients with RA or osteoarthritis (OA). The diagnosis of RA conformed to the 1987 revised American College of Rheumatology criteria (14). The postsurgical specimens were placed on ice and subsequently snap-frozen within 1 hr. Table 1 shows the clinical features of patients used in this study. All samples were obtained in accordance with the University of California at San Diego Institutional Review Board. Normal synoviocytes were obtained from autopsy specimens.
Table 1.
Clinical features of patients
| Feature | RA, n = 15 | OA, n = 6 |
|---|---|---|
| Age, yrs | 61 ± 3 | 68 ± 3 |
| Gender, M/F | 3/12 | 1/5 |
| Joint tissue, n | Knee (11), hand (3), shoulder (1), | Knee (5), hip (1) |
| Elbow (1) | ||
| Medications, n | NSAID (8), prednisone (8), | NSAID (1) |
| Methotrexate (7), sulfasalazine (1) | ||
| Hydroxychloroquine (1), | ||
| Azathioprin (2), previous methotrexate (2), | ||
| Previous alkylating agents (0) | ||
| Hematocrit, % | 37 ± 1 | 36 ± 1 |
| Platelet, ×103/dl | 330 ± 31 | 262 ± 30 |
| ESR, mm/hr | 30 ± 6 | N.D. |
n, number of patients in parentheses. N.D., not done; NSAID, nonsteroidal anti-inflammatory drug. Patient RA436 had two separate joint surgeries (shoulder and elbow).
Synoviocyte Culture.
Fibroblast-like synoviocytes were isolated by enzymatic dispersion of synovial tissues as previously described (15). Briefly, the tissues were minced and incubated with 1 mg/ml collagenase in serum-free DMEM (GIBCO/BRL) for 2 hr at 37°C, filtered through a nylon mesh, extensively washed, and cultured in DMEM supplemented with 10% fetal calf serum (GIBCO/BRL, endotoxin content <0.006 ng/ml), penicillin, streptomycin, and l-glutamine in a humidified 5% CO2 atmosphere. After overnight culture, nonadherent cells were removed, and adherent cells were cultivated in DMEM plus 10% fetal calf serum. At confluence, cells were trypsinized, split at a 1:3 ratio, and recultured in medium. Synoviocytes from passages 3–6 were used in these experiments, during which time they were a homogeneous population of fibroblast-like synoviocytes (<1% CD11b, <1% phagocytic, and <1% Fc-gamma RII receptor positive).
Preparation of RNA and RNA Mismatch Detection Assays (RMDAs).
Synovial tissue or skin from the juxta-articular incision was obtained at the time of joint replacement surgery and was snap-frozen in liquid nitrogen, and RNA was extracted using Trizol Reagent (GIBCO/BRL). Reverse transcription was performed using the Superscript Preamplification System (GIBCO/BRL) for first-strand cDNA synthesis, and then amplified using primers flanking p53 exons 4–11 (5′-ACCTA CCAGG GCAGC TACGG TTTC-3′; 5′-CCAGT CAGAT GGAGG GCGGT ATTT-3′). PCR amplification was performed using either Taq or Pfu polymerase, as indicated in the text. Wild-type p53 amplified from blood of a normal volunteer was used as a control template. A known mutant p53 template also was included as a mutant control. Nested primers containing T7 and SP6 phage promoters (5′-GATAA TACGA CTCAC TATAG GGCTT CTTGC ATTCT G-3′; 5′-TCATT TAGGT GACAC TATAG GACTT CAGGT GGCTG GA-3′) then were used to amplify exons 5–10 from all experimental samples. Products were checked, and RNA was transcribed using Mismatch Detect II (Ambion, Austin, TX) according to the manufacturer’s instructions, except that wild-type template and the SP6 and T7 series transcripts were generated separately and then hybridized to the appropriate wild-type transcript. Mismatches were detected by digestion with the supplied RNases, and the products were resolved on high-resolution agarose (Ambion). For second-round mismatch studies, the cDNA generated from the indicated tissues was subcloned into pCR-Script (Stratagene), and individual clones then were selected, linearized with BamHI, and used in RMDAs.
For detection of possible mutations in the retinoblastoma gene product, cDNA was prepared as above but then amplified with the following primers to examine two regions of RB1. For the region spanning bases 1,245–1,841, cDNA was amplified with 5′-ATTCC TCCAC ACACT CCAGT TAGG-3′ and 5′-ATCAG TTGGT CCTTC TCGGT CC-3′, followed by nested PCR with T7 and SP6 phage promoters (5′-GATAA TACGA CTCAC TATAG GTCCT CCACA CACTC CAGTT AGGAC-3′ and 5′-TCATT TAGGT GACAC TATAG GAGGT GAATC TGAGA GCCAT GCAAG-3′). For the region spanning bases 2,030–2,708, the corresponding sets of oligonucleotides were 5′-GCAAA TGCAG AGACA CAAGC AAC-3′, 5′-TCCTT CAGCA CTTCT TTTGA GCAC-3′, 5′-GATAA TACGA CTCAC TATAG GAAAG CAACC TCAGC CTTCC AGAC-3′, and 5′-TCATT TAGGT GACAC TATAG GACGG TCGCT GTTAC ATACC ATCTG-3′. In vitro transcription, hybridization to wild-type transcripts (prepared from normal blood), and RNase digestion then were performed as for the p53 mismatch detection.
Immunoprecipitation and Western Blotting.
For immunoprecipitation studies, tissue homogenates (100 mg) or cells (approximately 106 cells) were resuspended in cold PBS/0.1% sodium deoxycholate, frozen for 2 hr at −20°C, and thawed for 30 min on ice. After a second freeze-thaw cycle, the membranes were gently broken by mechanical pipetting. One milligram of protein in the lysate was precleared using 1 μg/ml of mouse IgG (Dako) and protein A/G agarose conjugate (Santa Cruz Biotechnology) for 1 hr on ice. The supernatant was immunoprecipitated using either PAb240 or DO1 at a final concentration of 1 μg/ml on ice. PAb240 detects a determinant on many mutant p53 proteins, but not on nondenatured wild-type p53 (16). DO1 binds to amino acids 21–25 on p53 and immunoprecipitates both mutant and wild-type protein (Calbiochem). After 1 hr, 10 μl of protein A/G agarose was added, and the solution was gently agitated overnight at 4°C. The precipitate was collected by centrifugation and washed 3 times with RIPA (PBS/1% Nonidet P-40/0.1% sodium deoxycholate/0.1% SDS). Immunoprecipitates were run on a 10% SDS/PAGE gel and transferred onto a nitrocellulose membrane at 140 mA in 25 mM Tris⋅Cl, pH 8.3/192 mM glycine/10% methanol. Filters were blocked with Tris-buffered saline plus 0.5% Tween-20 and 1.5% gelatin for 45 min. This was followed by incubation with biotinylated sheep polyclonal anti-p53 antibody (0.1 μg/ml) (Boehringer Mannheim) at 4°C overnight. The membrane was washed three times, and incubated with streptaviden-peroxidase (Boehringer Mannheim) at 1:5,000 dilution for 2 hr at room temperature. The proteins were visualized by chemiluminescence using hydrogen peroxide and luminol as a substrate (DuPont) using Kodak X-AR film.
RESULTS
RMDA on Synovial Tissue.
RA and OA synovial tissues were obtained at the time of joint replacement surgery, and RMDA was used to screen the synovial tissues for possible p53 mutations. Because synovial tissue is a mixture of many cell types (17) and any given p53 mutation might be relatively rare, we hybridized known amounts of mutant p53 RNA transcripts to wild-type transcripts to determine the relative concentration required for detection with this method. These studies indicated that at least 2–5% of the RNA pool must be mutant to be detected (data not shown).
As shown in Table 2 (first-round mismatch), transcripts prepared directly from RA synovia commonly contained mismatches indicating the possible presence of p53 mutations (see Fig. 1 for representative examples). To determine if p53 RNA mismatches were limited to the joint, skin from the juxta-articular incision was obtained from five RA patients at the time of joint surgery. Despite the presence of synovial p53 mismatches in each of these patients, mismatch bands were not detected in any extra-articular samples (see Table 2 and Fig. 1). Synovium and skin specimens were obtained from patients with OA at the time of joint replacement surgery and examined in the first-round mismatch assay, and no mismatches were found. Blood samples from two healthy volunteers and one RA patient (RA435) similarly contained no mismatches in p53 RNA (results not shown). The pattern of the RMDA varied for each of the synovial tissues analyzed, suggesting that the specific p53 sequence variants of each might differ. Also, the RMDAs from these tissues often had complex banding patterns consistent with multiple p53 mutations within an individual joint.
Table 2.
p53 mismatch analysis of arthritis tissue
| Patient | Synovium first-round mismatch | Synovium second-round mismatch, positive clones/total clones screened | Skin first-round mismatch | Skin second-round mismatch |
|---|---|---|---|---|
| RA429 | + | + | ||
| RA430 | + | 3/11 | ||
| RA431 | + | 4/8 | ||
| RA433 | + | 2/5 | ||
| RA435 | + | 3/8 | − | |
| RA436 | ||||
| Joint 1 | + | N.D. | − | |
| Joint 2 | + | 5/10 | N.D. | |
| RA444 | + | |||
| RA445 | + | 3/7 | − | 0/6 |
| RA448 | + | |||
| RA453 | + | 2/5 | − | 0/6 |
| RA456 | + | |||
| RA457 | ± | 2/5 | ||
| RA460 | + | − | 0/6 | |
| RA464 | − | |||
| RA467 | + | 1/3 | − | |
| OA320 | − | |||
| OA439 | − | − | ||
| OA440 | − | − | ||
| OA441 | − | − | ||
| OA442 | − | − | ||
| OA458 | − | − | 0/5 |
RNA was prepared from the indicated tissue, and RMDA was performed (see Methods). First-round mismatch refers to analysis of bulk cDNA (+, mismatch detected; −, no mismatch detected; ±, ambiguous result); second-round mismatch refers to assessment of clones derived from the bulk cDNA (numbers are mismatches/total clones analyzed). N.D., not determined.
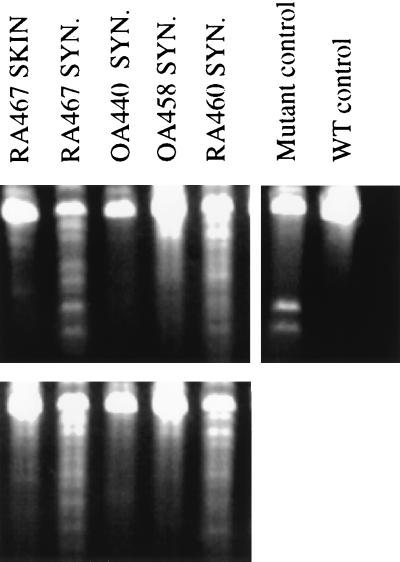
Figure 1.
RMDA for p53 from RA and OA tissue samples. RNA was prepared from skin or synovium (SYN) obtained from RA or OA patients, and RMDA was performed. A p53 mutant cDNA (Ambion) served as a control for positive mismatch, which appears as smaller fragments after RNase digestion. p53 from normal blood was used as wild-type template, and wild type hybridized to wild type served as a no-mismatch control. (Upper) RMDAs resulting from samples transcribed in the sense direction hybridized to wild-type antisense transcripts. RA467 SYN and RA460 SYN lanes demonstrated intense complex banding patterns, suggesting the presence of multiple mutations. RA467 SKIN, OA440 SYN, and OA458 SYN showed only very faint nonspecific bands. (Lower) The converse hybridizations with wild-type sense and sample antisense transcripts, with similar results.
Synovial transcripts from one RA patient (RA435, joint 2) with documented p53 synovial mismatch were subjected to RMDA for the retinoblastoma gene RB1 (see Methods) and no mismatch was found. This suggests that the observed mismatches in p53 transcripts are not necessarily a function of a general increase in somatic mutations, but instead may represent a selectable event.
To control for possible errors introduced during PCR amplification, some of the samples were analyzed after amplification with Pfu DNA polymerase, which has a significantly higher fidelity than Taq polymerase (18). These included two synovial samples (RA429 and RA446) and one blood sample (RA446). Mismatches were observed only in the synovial samples.
RMDA on p53 cDNA Subclones Isolated from RA Synovial Tissue.
To determine if first-round mismatches were a consequence of mutant p53 sequences, the product of the initial reverse transcription–PCR on synovial tissue or skin was subcloned and re-evaluated by RMDA. Because single clones were studied in the second round, sensitivity issues due to a mixture of normal and mutant p53 faced in the first-round mismatch assay were not encountered. The cDNA from synovia of several RA patients were studied in this manner, and 40 ± 2% of individual synovial subclones were positive. The ethidium-stained gels again showed a variety of mismatch patterns, suggesting more than one mutation within a single joint. As a control, skin samples from three RA patients (including RA445 and RA453, which had positive first- and second-round mismatches in synovial transcripts) and 1 OA patient (OA458) were examined by similar subcloning and RMDA despite having negative first-round results. Second-round mismatches were not found in any of the subclones from these skin samples. Thus, second-round mismatch detection assays suggest a surprisingly high prevalence of mutant p53 transcripts in RA synovium.
In two cases both the first- and second-round RMDA (RA429 synovium, RA446 synovium), and in one case the second-round RMDA (RA430), used Pfu rather than Taq polymerase (18) (see above). The identification and frequency of mismatches in these cases was similar to that of the others. Together with the failure to detect mismatches in p53 transcripts isolated from other tissues (Table 2), these observations suggest that PCR artifacts introduced by Taq are unlikely to account for the mismatches observed in synovial p53 transcripts.
Sequence Analysis of Synovial Tissue p53 cDNA Subclones.
Individual RA synovial subclones with mismatches then were sequenced. When clones were negative in the second-round assay, the sequence was always wild type (results not shown). A variety of missense and silent mutations were identified among the individual subclones that were sequenced. Table 3 shows the data for all sequenced clones. Virtually all of the mutations were different, and as many as three distinct mutations were isolated from a single joint. In one case (RA435), several clones with an identical mutation were observed, although this was silent. The skin of this patient had a normal first-round mismatch, indicating that the conserved mutation was specific to the synovium. Point mutations and deletions were randomly distributed throughout exons 5–10. Some individual clones contained two separate mutations (RA445 and RA453). The presence of several different mutations in transcripts from a single joint indicates that these are not clonally derived, but instead may be oligoclonal, analogous to the different p53 mutations observed in actinic keratosis samples from a single patient (19).
Table 3.
P53 mutations identified from RA synovial tissues
| Patient | Predicted amino acid at mutation site | Nucleotide sequence change | Base change |
|---|---|---|---|
| RA429 | R333H | CGT > CAT | G > A |
| N239S | AAC > AGC | A > G | |
| L369fs | G deletion, base 1028 | G deletion | |
| RA430 | V203V (conserved) | GTG > GTA | G > A |
| I232M | ATC > ATG | C > G | |
| RA435 | L188L (conserved) | CTG > CTA | G > A |
| RA436 | E224fs | G deletion, base 791 | G deletion |
| RA445 | Two mutations in a single clone: R174G; G199G | AGG > GGG and GGA > GGG | A > G and A > G |
| RA453 | Two mutations in a single clone: K139R; H193Y | AAG > AGG and CAT > TAT | A > G and C >T |
| RA457 | L257E | CTG > CAG | T > A |
All of the subclones that were sequenced from the second-round mismatch analyses from Table 2 are presented. fs, frame shift.
In addition to sequencing clones with mismatches, we sequenced two clones generated after PCR amplification of a wild-type p53 cDNA and two others amplified from blood transcripts. The full-length sequences in these cases were completely wild type, indicating that errors introduced by PCR amplification are relatively rare and cannot account for the substitutions we observed in the synovial transcripts.
RMDA and Sequence Analysis of Fibroblast-Like Synoviocyte p53 cDNA.
Because our original observation suggested an abundance of DNA strand breaks and p53 overexpression in cultured synovial lining fibroblasts, we determined whether these cells might harbor p53 mutations. Fibroblast-like synoviocytes were obtained from enzymatically dispersed synovial tissue and studied at passage 3 or 4 when they are a homogenous population of fibroblast-like cells (15). Cultured synoviocytes are thought to be derived from type B synoviocytes in the synovial intimal lining (1–3). Four RA cell lines were examined, and two were positive in the first-round mismatch assay. Second-round mismatches were found in 25–40% of cDNA clones from these two lines, and sequence analysis revealed missense mutations (Table 4). In one case, both a cell line and the original tissue were examined; although both had mutations, the specific mutations were different. This is not surprising, because multiple mutations were found in individual joints (see above). Nevertheless, the presence of mutations in these cell lines suggests that the mutations found in the joints are in synoviocytes.
Table 4.
Mutations in RA synoviocyte lines
| Patient | First-round mismatch | Second-round mismatch | Predicted amino acid at mutation site | Nucleotide sequence | Base change |
|---|---|---|---|---|---|
| RA455 | − | ||||
| RA456 | + | 8/20 | C277C (silent); C176R; R213* | TGT > TGC Two mutations in a single clone: TGC > CGC and CGA > TGA | T > C T > C and C > T |
| RA457 | + | 4/16 | E258E (silent); S227P; Y327Y (silent) | GAA > GAG Two mutations in a single clone: TCT > CCT and TAT > TAC | A > G T > C and T > C |
| RA459 | − |
Synoviocytes from the joints of the indicated patients were cultured. After three or four passages synoviocytes were assessed by RMDA for p53 mutations. First- and second-round mismatch analysis was performed as in Table 2.
Stop codon.
Analysis of p53 Mutations in RA.
Fourteen of the 18 p53 mutations observed in synovium or synoviocyte lines were transitions in the sense or antisense strands (G→A or A→G; T→C or C→T) (see Table 4), consistent with a common mechanism of mutation, such as oxidative deamination by NO or oxygen radicals (20, 21). In addition, two G deletions and two transversions (T→A and C→G) were observed. Many of the predicted amino acid substitutions are identical or at the same position as those observed in a variety of tumor types (22), suggesting that these may have occurred at known “hot spots.”
Detection of Immunoreactive Mutant p53 Protein in RA Synovium.
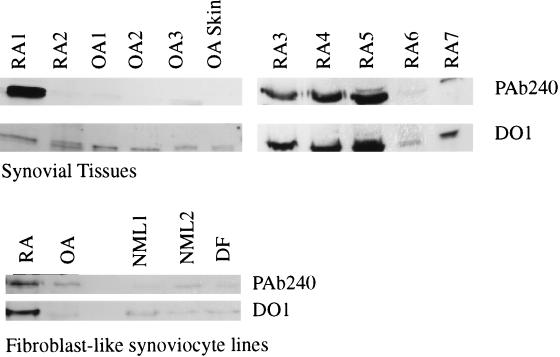
We previously have shown that p53 protein is expressed in the RA synovial lining using immunohistochemistry and the mAb PAb1801, which detects wild-type and mutant p53 protein (23, 24). This observation was confirmed and extended in the present studies using the mAb PAb240, which detects a determinant on many mutant p53 proteins, but not on nondenatured wild-type p53 (16). PAb240 was used in immunoprecipitation experiments to confirm the presence of abnormal p53 protein (see Fig. 2). Immunoreactive p53 protein was detected in OA and RA samples using the mAb DO1, which precipitates both mutant and wild-type p53, whereas normal synoviocytes and dermal fibroblasts contained little or no p53 protein. However, when PAb240 was used as the precipitating antibody, the OA synovial tissues, OA synoviocytes, OA skin samples, normal dermal fibroblasts, and normal synoviocytes contained little or no immunoreactive product. An RA fibroblast-like synoviocyte line as well as most RA synovial tissues studied (4/7) contained abundant PAb240-reactive protein. These data indicate that the p53 protein in the non-RA samples is wild type, whereas mutant p53 protein is present in RA synoviocytes and synovium.
Figure 2.
p53 protein detection by immunoprecipitation. p53 protein expression was analyzed in synovial tissue (RA and OA), skin (OA), fibroblast-like synoviocytes [RA, OA, and normal (NML)], and dermal fibroblasts (DF). Different samples are from different patients and are numbered arbitrarily. Immunoprecipitation was performed as described in Methods using either PAb240 (detects mutant but not wild-type p53) or DO1 (detects mutant and wild-type p53). Note the discordant expression of DO1 and PAb240 precipitable protein in OA joint samples, whereas abundant p53 was detected in RA joint tissue using PAb240.
DISCUSSION
RA evolves from a local inflammatory disease to a chronic process with distinct inflammatory and destructive components. Accumulating evidence suggests that this transition might be due to partial transformation of RA fibroblast-like synoviocytes. For instance, RA synoviocytes can proliferate in an anchorage-independent manner and lack contact inhibition (5–7). In addition, cultured synovial fibroblasts that have been coimplanted into severe combined immunodeficient mice with cartilage explants autonomously invade into the cartilage matrix, whereas OA synoviocytes and normal dermal fibroblasts do not (25). These data provide evidence that synoviocytes are irreversibly altered or imprinted in RA and that they remain activated even after removal from the articular inflammatory milieu.
In the course of studying synoviocyte function and transformation in RA, we and others recently observed that DNA strand breaks are abundant in the RA synovial intimal lining (8, 9). This process is likely a result of the toxic effects of locally produced NO and oxygen radicals. Despite the excessive DNA fragmentation, only rare cells exhibit morphologic evidence of apoptosis (8, 9). This discrepancy implies a defect in the ability to delete damaged synovial cells and led us to examine p53 as a potential key regulator of DNA repair, cell replication, and apoptosis in RA. These studies demonstrated marked overexpression of p53 protein in rheumatoid synovium and in resting RA fibroblast-like synoviocytes (10).
If DNA strand breaks in RA synovium do not necessarily lead to cell deletion, then synoviocytes could survive longer than normally anticipated and accumulate in the intimal lining. This hypothesis stimulated studies to investigate the possibility that the p53 gene in the RA joint is abnormal. Using an RMDA, RA synovial cDNA, but not matched skin or blood samples from patients, showed evidence of p53 mutations. In addition, synovium and skin from patients with OA did not have p53 mismatches. Subsequent subcloning and sequence analysis demonstrated that 40% of p53 cDNA isolated from RA synovium had mutations. This relatively high frequency of mutations in synovial transcripts does not imply that a similar high frequency of cells in the joint have mutations, because a minority of cells might produce a disproportionally large fraction of the p53 mRNA.
The specific types of base changes identified in p53 mutations can be a clue to the mutagenic stimulus. Exposure to ultraviolet light, for example, is correlated with transition mutations at dipyrimidine sites (26). In contrast, aflatoxin B1 exposure results in G/T transversions that lead to amino acid substitutions as seen in hepatocellular carcinoma (27); cigarette smoke also leads to base transversions (28). Furthermore, one DNA strand can be more susceptible than the other; the predominance of C to T transitions in skin carcinomas suggests that the nontranscribed strand is more likely to be the site of mutation (29). Greater than 80% of the mutations we identified in synovium and cultured synoviocytes are G/A and T/C transitions. Such mutations are characteristic of oxidative deamination by NO or oxygen radicals (20, 21). Similar mutations occur in colon carcinoma and are thought to be due to endogenous NO production (30). This supports our hypothesis that a genotoxic local environment in the chronically inflamed synovium accounts for mutations in the p53 gene in RA.
A number of caveats should be recognized when interpreting these findings. First, one must always be aware of potential artifacts, especially with PCR. However, mismatches and sequence abnormalities were found only in RA synovium, which argues strongly against this explanation and suggests that the abnormalities are due to somatic mutations. In addition, the consistent finding of transition mutations implies a specific mutagenic stimulus instead of a random artifact. Finally, amplification with Pfu rather than Taq polymerase in several cases had no effect on either the identification or frequency of mismatches detected in the RMDA. Thus, PCR artifacts due to the use of Taq polymerase are unlikely to have significantly contributed to our findings.
A second issue is whether the drugs used to treat arthritis could contribute to our findings. Notably, none of the patients had been treated with alkylating agents. The lack of mutations in other tissues of the RA patients and the high percentage of transition mutations also argues against drug-induced mutations.
It is important to recognize that the p53 mutations do not cause RA; rather, they are probably the result of intense local chronic inflammation. We propose that a burden of mutations accumulates over time and that specific alterations in the p53 gene (or perhaps other genes that regulate apoptosis or the cell cycle) can contribute to the autonomy of synoviocytes and perpetuation of disease. For instance, excessive interleukin-6 production by fibroblast-like synoviocytes could result from a failure of mutant p53 to repress transcription, because p53 protein is a potent repressor of the interleukin-6 promoter (31). Some of the mutations identified in the p53 gene reside in previously identified hot spots that control p53 binding to DNA or other aspects of p53 action, and thus it is likely that the abnormal proteins are functionally inactive. Several of the mutations or sites of mutations identified in RA synovium are known to be associated with neoplastic diseases (22).
Because RA is a polyarticular disease, local mutations likely occur in multiple joints independently. Also, single joints can exhibit multiple mutations, but no evidence shows that a single synovial tissue has clonal expansion of an individual clone. Rather, multiple foci probably occur, with local expansion. A similar process operates in sun-exposed skin where chronic low-dose ultraviolet irradiation leads to actinic keratoses, associated with multiple p53 mutations (32). Such p53 mutations do not transform this tissue; rather, it is subsequent mutagenic exposure that leads to a “second hit,” which contributes to malignancy. More recently, it has been shown that normal skin can harbor clones of cells carrying p53 mutations (33). Similarly, the RA synovium is chronically exposed to genotoxic insults, such as NO and oxygen radicals that can produce transition mutations by oxidative deamination (34–36).
The process whereby chronic inflammation promotes local p53 mutations is probably not unique to RA, but might be a common feature of many diseases. One example is ulcerative colitis, which is marked by long-standing inflammation, p53 mutations, and malignancy (37, 38). Why p53 mutations in synoviocytes do not lead to malignant tumors in RA is not clear. However, the synovium is uniquely resistant to transformation. Synovial sarcomas are exceedingly rare; when they do occur they arise from the supporting joint structures rather than from the synovial lining and have the ultrastructural appearance of epithelial cells instead of mesenchymal cells (39). Finally, p53 is not an oncogene, and mutations that contribute to malignant transformation in other tissues may not affect synoviocytes in the same way.
The observation that p53 mutations occur in RA synovium and that similar mutations are present in cultured synoviocytes suggests a molecular mechanism for many aspects of synoviocyte biology. For instance, we recently have found that suppression of p53 in cultured synoviocytes with the papilloma virus protein E6 protein significantly increases their growth rate and decrease susceptibility to apoptosis (K. R. Aupperle, D. L. Boyle, M. Hendrix, N.J.Z., M. Barbosa, and G.S.F., unpublished observations). Additional studies are in progress to characterize the activity of specific RA p53 mutations and determine functional consequences of p53 suppression in cultured synoviocytes.
The notion that somatic mutations occur in apoptosis and cell cycle regulating genes in RA supports the hypothesis that chronic RA has a transformed, invasive component and suggests that novel therapeutic strategies might be applicable to arthritis and perhaps other inflammatory diseases (40). Such approaches might have the advantage of directly addressing tissue destruction and altering the natural history of the disease rather than simply suppressing inflammation.
Acknowledgments
This work was supported by grants from the National Institutes of Health to G.S.F., N.J.Z., and D.R.G.
ABBREVIATIONS
- RA
rheumatoid arthritis
- OA
osteoarthritis
- RMDA
RNA mismatch detection assay
References
- 1.Firestein G S. Arthritis Rheum. 1996;39:1781–1790. doi: 10.1002/art.1780391103. [DOI] [PubMed] [Google Scholar]
- 2.Pitsillides A A, Wilkinson L S, Mehdizadeh S, Bayliss M T, Edwards J C. Int J Exp Pathol. 1993;74:27–34. [PMC free article] [PubMed] [Google Scholar]
- 3.Morales-Ducret J, Wayner E, Elices M J, Alvaro-Gracia J M, Zvaifler N J, Firestein G S. J Immunol. 1992;149:1424–1431. [PubMed] [Google Scholar]
- 4.Gay, S., Gay, R. E. & Koopman, W. J. (1993) Ann. Rheum. Dis. 52, Suppl. 1, S39–S47. [DOI] [PMC free article] [PubMed]
- 5.Lafyatis R, Remmers E F, Roberts A B, Yocum D E, Sporn M B, Wilder R L. J Clin Invest. 1989;83:1267–1276. doi: 10.1172/JCI114011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucala R, Ritchlin C, Winchester R, Cerami A. J Exp Med. 1991;173:569–574. doi: 10.1084/jem.173.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remmers E F, Lafyatis R, Kumkumian G K, Case J P, Roberts A B, Sporn M B, Wilder R L. Growth Factors. 1990;2:179–188. [PubMed] [Google Scholar]
- 8.Firestein G S, Yeo M, Zvaifler N J. J Clin Invest. 1995;96:1637–1638. doi: 10.1172/JCI118202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima T, Aono H, Hasunuma T, Yamamoto K, Shirai T, Hirohata K, Nishioka K. Arthritis Rheum. 1995;38:485–491. doi: 10.1002/art.1780380405. [DOI] [PubMed] [Google Scholar]
- 10.Firestein G S, Nguyen K, Aupperle K, Yeo M, Boyle D L, Zvaifler N J. Am J Pathol. 1996;149:2143–2151. [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigues N R, Rowan A, Smith M E, Kerr I B, Bodmer W F, Gannon J V, Lane D P. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar G, Yoon H A, Sommer S S. Genomics. 1992;13:441–443. doi: 10.1016/0888-7543(92)90266-u. [DOI] [PubMed] [Google Scholar]
- 13.Davidoff A M, Humphrey P A, Iglehart J D, Marks J R. Proc Natl Acad Sci USA. 1991;88:5006–5010. doi: 10.1073/pnas.88.11.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnett F C, Edworthy S M, Bloch D A, McShane D J, Fries J F, Cooper N S, Healey L A, Kaplan S R, Liang M H, Luthra H S, Medsger T A, Jr, Mitchell D M, Neustadt D H, Pinals R S, Schaller J G, Sharp J T, Wilder R L, Hunder G G. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 15.Alvaro-Gracia J M, Zvaifler N J, Firestein G S. J Clin Invest. 1990;86:1781–1790. doi: 10.1172/JCI114908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gannon J V, Greaves R, Iggo R, Lane D P. EMBO J. 1990;9:1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firestein G S. In: Rheumatoid Synovitis and Pannus. Klippel J, Dieppe P, editors. St. Louis: Mosby; 1994. pp. 3.12.1–3.12.30. [Google Scholar]
- 18.Cline J, Braman J C, Hogrefe H H. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson M A, Einspahr J G, Alberts D S, Balfour C A, Wymer J A, Welch K L, Salasche S J, Bangert J L, Grogan T M, Bozzo P O. Cancer Lett. 1994;85:23–29. doi: 10.1016/0304-3835(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 20.Wink D A, Kasprzak K S, Elespuru C M, Misra M, Dunams T M, Cebula T A, Koch W H, Andrews A W, Allen J S, Keefer L K. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen T, Brunson D, Crespi C L, Penman B W, Wishnok J S, Tannenbaum S R. Proc Natl Acad Sci USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroud C, Verdier F, Soussi T. Nucleic Acids Res. 1996;24:147–150. doi: 10.1093/nar/24.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vojtesek B, Bartek J, Midgeley C A, Lane D P. J Immunol Methods. 1992;151:237–244. doi: 10.1016/0022-1759(92)90122-a. [DOI] [PubMed] [Google Scholar]
- 24.Banks L, Matlashewski G, Crawford L. Eur J Biochem. 1986;159:529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- 25.Muller-Ladner U, Kriegsmann J, Franklin B N, Mastumoto S, Geiler T, Gay R E, Gay S. Am J Pathol. 1996;149:1607–1615. [PMC free article] [PubMed] [Google Scholar]
- 26.Brash D E, Rudolph J A, Simon J A, Lin A, McKenna G J, Baden H P, Halperin A J, Ponten J. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu I C, Metcalf R A, Sun T, Welsh J A, Wang N J, Harris C C. Nature (London) 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 28.Takeshima Y, Seyama T, Bennett W P, Akiyama M, Tokuoka S, Inai K, Mabuchi K, Land C E, Harris C C. Lancet. 1993;342:1520–1521. doi: 10.1016/s0140-6736(05)80087-x. [DOI] [PubMed] [Google Scholar]
- 29.Bohr V A. Carcinogenesis. 1991;12:1983–1992. doi: 10.1093/carcin/12.11.1983. [DOI] [PubMed] [Google Scholar]
- 30.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 31.Santhanam U, Ray A, Sehgal P B. Proc Natl Acad Sci USA. 1991;88:7605–7609. doi: 10.1073/pnas.88.17.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler A, Jonason A S, Leffell D J, Simon J A, Sharma H W, Kimmelman J, Remington L, Jacks T, Brash D E. Nature (London) 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 33.Jonason A S, Kunala H, Price G J. Proc Natl Acad Sci USA. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh D, Nazhat N B, Fairburn K, Sahinoglu T, Blake D R, Jones P. Ann Rheum Dis. 1995;54:94–99. doi: 10.1136/ard.54.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grootveld M, Henderson E B, Farrell A, Parkes D R, Haycock P. Biochem J. 1991;273:459–467. doi: 10.1042/bj2730459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrell A J, Blake D R, Palmer R M, Moncada S. Ann Rheum Dis. 1992;51:1219–1222. doi: 10.1136/ard.51.11.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brentnall T A, Crispin D A, Rabinovitch P S, Haggitt R C, Rubin C E, Stevens A C, Burner G C. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 38.Harris C C. Toxicology Lett. 1995;82–83:1–7. doi: 10.1016/0378-4274(95)03643-1. [DOI] [PubMed] [Google Scholar]
- 39.Dickersin G R. Ultrastruct Pathol. 1991;15:379–402. doi: 10.3109/01913129109016247. [DOI] [PubMed] [Google Scholar]
- 40.Bischoff J R, Kim D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J, Sampson-Johannes A, Fatteay A, McCormick F. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]