Abstract
PML/RARα is the abnormal protein product generated by the acute promyelocytic leukemia-specific t(15;17). Expression of PML/RARα in hematopoietic precursor cell lines induces block of differentiation and promotes survival. We report here that PML/RARα has a potent growth inhibitory effect on all nonhematopoietic cell lines and on the majority of the hematopoietic cell lines tested. Inducible expression of PML/RARα in fibroblasts demonstrated that the basis for the growth suppression is induction of cell death. Deletion of relevant promyelocytic leukemia (PML) and retinoic acid receptor (RARα) domains within the fusion protein revealed that its growth inhibitory effect depends on the integrity of the PML aminoterminal region (RING, B1, B2, and coiled coil regions) and the RARα DNA binding region. Analysis of the nuclear localization of the same PML/RARα deletion mutants by immunofluorescence and cell fractionation revealed that the biological activity of the fusion protein correlates with its microspeckled localization and its association to the nuclear matrix. The PML aminoterminal region, but not the RARα zinc fingers, is required for the proper nuclear localization of PML/RARα. We propose that the matrix-associated microspeckles are the active sites of PML/RARα and that targeting of RARα sequences to this specific nuclear subdomain through PML sequences is crucial to the activity of the fusion protein on survival regulation.
Acute promyelocytic leukemia (APL) genetically is characterized by the PML/RARα fusion gene that is formed as a consequence of the 15;17 translocation, whose breakpoints are located within the promyelocytic leukemia (PML) locus on chromosome 15 and the retinoic acid receptor (RARα) locus on chromosome 17. The PML/RARα gene codes for a fusion protein that retains most of the functional domains of the parental PML and RARα proteins (1, 2).
Indirect evidence suggests that this genetic recombination is crucially involved in the pathogenesis of the disease: (i) the t(15;17) is present in 90% of APL cases and frequently is the only cytogenetic aberration; (ii) the chimeric PML/RARα gene also is detected in cytogenetically negative APL cases; and (iii) unlike fusion genes in other acute leukemias, detection of the PML/RARα transcripts after anti-leukemic treatment is invariably correlated with disease relapse (1, 2). Recent support for the pathogenetic role of the PML/RARα recombination comes from the finding that expression of PML/RARα in the chicken and mouse bone marrow results in alterations of the hematopoietic myeloid development with typical APL phenotypic features (3–5).
Little is known of the biological and molecular mechanisms of the leukemogenetic activity of PML/RARα. Its expression in hematopoietic precursor cells blocks terminal differentiation and promotes survival (6–8). Because these two biological activities correlate with prominent phenotypic features of APL, that is accumulation of hematopoietic precursors blocked at the promyelocytic stage, they may reflect the in vivo leukemogenetic activity of PML/RARα.
The PML aminoterminal portion retained in the fusion protein contains a number of characterized motifs including a zinc binding domain called RING finger, two newly defined cysteine/histidine-rich protein motifs called B-boxes (B1 and B2), and a coiled coil region (9, 10). The function of the RING and B-box regions is unknown, whereas the coiled coil is a dimerization interface, through which PML/RARα forms homodimers and heterodimers with PML (11). PML/RARα retains the RARα DNA and retinoic acid (RA) binding domains and the retinoid receptor (RXR) dimerization interface. Accordingly, the fusion protein is able to transactivate in vitro RA target genes (4–7) and to bind RXRs (11) and RA (12). Functional analysis of a variety of PML/RARα mutants revealed that the PML coiled coil region and the RARα DNA binding domains are essential for the capacity of PML/RARα to block differentiation and that they only exert their functions within the context of the fusion protein (13).
We investigated the effects of PML/RARα on survival and its mechanisms of action. We report that PML/RARα induces cell death in all nonhematopoietic and in most of hematopoietic cell lines tested. This effect depends on the localization of the fusion protein to nuclear matrix-associated microspeckles and on both PML (RING, B-boxes, and coiled coil regions) and RARα (DNA binding region) sequences.
MATERIALS AND METHODS
Cell Culture and Western Blotting.
The various cell lines were cultured in DMEM supplemented with 10% bovine calf serum. Transfections were performed by the calcium phosphate precipitation method. PML/RARα expression was induced by 100 μM ZnSO4 (Zn) or 1 mM 17βE. Western blot analysis were performed as described (13) using an anti-RARα serum against the F domain (13) and revealed with the ECL (Amersham) method.
Cloning and Sequence of Spontaneous PML/RARα Mutants.
PA317 clones expressing rearranged PML/RARα proteins were identified by Western blotting. The coding region of the mutated PML/RARα cDNA was isolated from PA317 genomic DNA by PCR and sequenced by the Sanger dideoxy-mediated chain termination method.
Colony Formation Assay.
NIH 3T3 or U2OS (5 × 105) cells were plated in a 10-cm dish and transfected with variable amount of expression vectors. Three days later, cells were split 1:10 or 1:5 and subcultured into medium containing 500 mg/ml G418. In selected experiments 10−6 M all-trans RA was added 1 day after transfection and the RA-containing medium changed every 4 days. Drug-resistant colonies were counted after crystal violet staining.
Growth Curves and Cell Death Evaluation.
Cells were plated in 6-well dishes at a concentration of 1 × 105 cells per well and grown for 96–120 hr in the presence or absence of 100 μM Zn or 1 mM 17βE. Cell proliferation was evaluated each day by direct cell counting using the trypan blue exclusion method. Cell death evaluation was performed analyzing the DNA content and cell cycle distribution by flow cytometric analysis of propidium iodide (PI)-stained cells (14) and by gel electrophoresis of genomic DNA.
Immunofluorescence Staining and Microscope Analysis.
Adherent growing cells were fixed for 10 min in 100% methanol at room temperature and then permeabilized for 2 min in acetone at −20°C. PML staining was performed with two different antibodies: the PGM-3 mAb, directed against the PML N-terminal region (15), and PG serum, against the PML α-helix region (a gift from P. Greer, Queen’s University, Kingston, Ontario). RARα staining was performed with a rabbit polyclonal antibody directed against the F-domain of the protein.
Cell Fractionation Experiments.
Cells (30 × 106) were pelletted and washed with PBS, resuspended in 3 packed cell volume of buffer A (10 mM Hepes, pH 8/1,5 mM MgCl2/10 mM KCl/1 mM DTT), left swelling on ice for 15 min, and lysed with a gauge needle. Homogenates were spun for 20 sec: the supernatant was collected as cytoplasm. The pellet was resuspended in 1 packed cell volume of buffer C (20 mM Hepes, pH 8/1,5 mM MgCl2/25% glycerol/420 mM NaCl/0,2 mM EDTA, pH 8/1 mM DTT/0,5 mM phenylmethylsulfonyl fluoride) and incubated on ice for 60 min. Nuclear debris were spun, resuspended in 100 ml × 30 × 106 cells SDS/PAGE loading buffer (50 mM Tris⋅Cl, pH 6, 8/100 mM DTT/2% SDS/0,1% bromophenol blue/10% glycerol), sonicated, and considered the matrix fraction, whereas the supernatant was considered the soluble nucleoplasm and chromatin (27).
RESULTS
Selective Permissivity of Various Cell Lines for PML/RARα Expression.
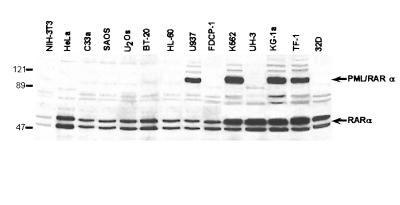
To investigate the effect of PML/RARα on survival, we transduced the fusion protein cDNA into a variety of cell lines of both hematopoietic (HL60, U937, K562, TF-1, FDCP-1, 32D, KG-1a, and Epstein–Barr virus-transformed lymphoblastoid UH3 cells) and nonhematopoietic (NIH 3T3 and PA317 fibroblasts, HeLa and C33a cervix carcinoma, U2OS and SAOS osteosarcoma, and BT-20 breast carcinoma cells) origin. The PML/RARα cDNA cloned into the LXSN retroviral vector (LPRSN, ref. 13) was transfected or electroporated in all of the listed cell lines. Mass cell populations were selected and analyzed by Western blotting. PML/RARα expression was detected in a subset of hematopoietic cell lines of myeloid leukemia origin (K562, U937, TF-1, and KG-1a), but in none of the nonhematopoietic cell lines (Fig. 1; see also ref. 13).
Figure 1.
Selective permissivity of hematopoietic and nonhematopoietic cell lines for PML/RARα expression. Western blot analysis of PML/RARα expression of different cell lines transduced with the LPRSN expression vector. Molecular weight markers are given on the left of the blot.
PML/RARα expression then was analyzed in a series of cell clones derived from independent mass cultures of cells that did not express PML/RARα (HL-60, 32D, FDCP Mix-A4, and PA317). No expression was detected in any of the 51 HL-60, 15 32D, and 12 FDCP clones analyzed and in 40 of the 64 PA317 clones analyzed (data not shown). Of the remaining 24 PA317 clones, 15 expressed low levels of the fusion protein and nine expressed high levels of anti-RARα-reacting polypeptides with molecular masses between 70 and 80 kDa (not shown).
To determine whether the lack of expression was an intrinsic property of some cell lines or the consequence of their selection in the presence of PML/RARα expression, we analyzed fusion protein expression after transient transfection and pharmacological selection. As demonstrated by anti-PML immunostaining, approximately 15% 32D cells displayed the typical microspeckled nuclear localization pattern of PML/RARα 48 hr after electroporation (not shown). However, after G418 selection of the same transfected population, approximately 5% cells still expressed the exogenous protein, but in a nonmicrospeckled pattern (not shown). Because the anti-PML PG-M3 antibody used does not react with mouse PML proteins (15), the exogenous protein alone is responsible for the anti-PML labeling in 32D cells. We interpret this change in localization as a consequence of mutations within the PML/RARα coding sequence due to negative selection against PML/RARα expression.
Permissivity for PML/RARα expression seems to differ among cell lines and expression of an intact PML/RARα protein appears to be counterselected during the growth of the majority of the cell lines tested. Only a few cell lines of hematopoietic derivation (KG-1a, U937, K562, and TF-1) were permissive for PML/RARα expression.
PML/RARα Inhibits Fibroblast Growth.
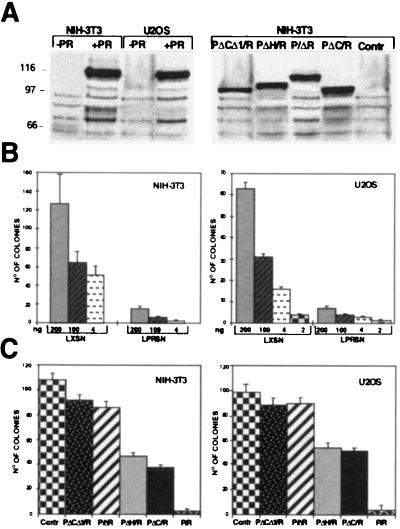
To investigate the mechanism underlying the variable permissivity for PML/RARα expression, we tested the capacity of nonpermissive cells to form G418-resistant colonies in the presence of PML/RARα expression (colony formation assay). For this purpose, NIH 3T3 and U2OS cells were transfected with the LPRSN or the empty LXSN expression vectors, and, after 20 days of G418 selection, stained with crystal violet and evaluated for the number of resistant colonies. A dramatic drop in the capacity of both NIH 3T3 (Fig. 2B, Left) and U2OS (Fig. 2B, Right) cells to form colonies in the presence of PML/RARα expression resulted. The level of PML/RARα expression 48 hr after transfection was high (Fig. 2A, Left), but became very low or absent after selection, as evaluated by Western blotting of the rare resistant colonies (not shown).
Figure 2.
Effects of PML/RARα expression on growth of NIH 3T3 and U2OS cells. (A) Western blot analysis of PML/RARα (PR) and mutant protein expression. NIH 3T3 or U2OS cells were transfected with the indicated expression vectors and analyzed 48 hr after by Western blotting using anti-RARα antibodies. Control lanes (Contr) are cells transfected with the empty vector. (B and C) Colony formation assays. NIH 3T3 (Left) or U2OS (Right) were transfected with the indicated amounts of the PML/RARα expression vector (B) or with 1 μg of the various deletion mutant fusion proteins expression vectors (C) and then G-418 selected. Resistant colonies were counted after crystal violet staining. Results are given as the median of three independent triplicate experiments.
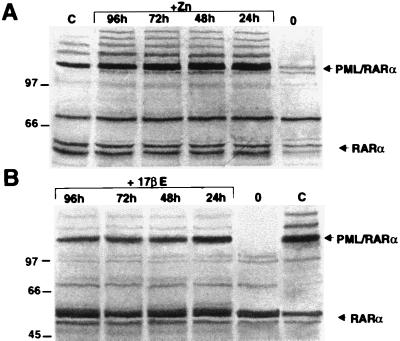
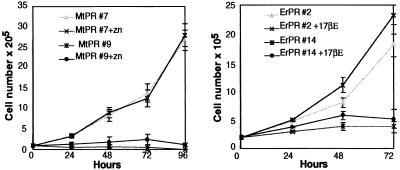
To investigate the cellular mechanism responsible for this growth inhibition, we analyzed the effect of inducible PML/RARα expression in nonpermissive cells using two different inducible expression systems. In the first, PML/RARα was cloned downstream from a minimal promoter containing four Gal-4 binding sites (pG-GPT) (gift from S. Braselmann, IMP, Research Institute of Molecular Pathology, Wien, Austria). The resulting expression vector (pGPR-GPT) was cotransfected into NIH 3T3 cells with a second expression vector that codes for a Gal4-estrogen receptor fusion protein (pMV-GalEr-vp). Transcription from the minimal promoter is activated by the Gal4-estrogen fusion in the presence of 17βE. In the second expression system, PML/RARα was cloned under the control of the Zn-inducible mouse metallothionene-1 (MoMt-1) promoter and the resulting expression vector (MtPR) transfected into NIH 3T3 cells. Clones with inducible PML/RARα expression were identified by Western blotting. PML/RARα expression was low or undetectable in the absence of Zn (NIH-MtPR clone 7, Fig. 3A) or 17βE (NIH-ErPR clone 2, Fig. 3B) induction, whereas it was significant after 24 hr of induction and remained stable for 4 days (Fig. 3 A and B). The effect of PML/RARα-inducible expression on the growth potential of NIH 3T3 cells next was examined in two Zn- and two 17βE-inducible clones (NIH-MtPR 7 and 9, and NIH-ErPR 2 and 14, respectively). Two clones each transfected with the Mt and pG-GPT empty vectors (NIH-Mt 2 and 4, and NIH-Er 1 and 3, respectively) served as controls. Cell growth was monitored by the trypan blue exclusion test. Control cells grew exponentially, and neither Zn or 17βE treatment had significant effects on cell number (not shown). Addition of Zn to NIH-MtPR or 17βE to NIH-ErPR clones almost completely prevented the expansion of the cell population (Fig. 4).
Figure 3.
Western blot analysis of PML/RARα-inducible expression. (A) Time course of PML/RARα induction upon 100 μM Zn treatment in the NIH-MtPR7 cells. (B) Time course of PML/RARα induction upon 1 mM 17βE treatment in the NIH-ErPR2 cells.
Figure 4.
Growth inhibition of NIH 3T3 fibroblasts by inducible PML/RARα expression.
PML/RARα Induces Cell Death of Fibroblasts.
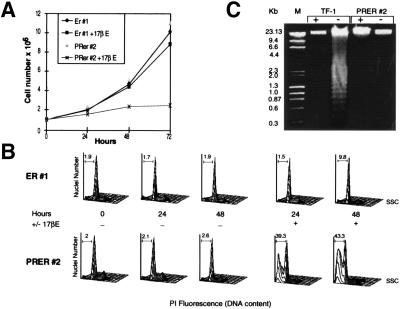
Trypan blue staining of Zn-treated NIH-MtPR and 17βE-treated NIH-ErPR cells revealed increased numbers of dead cells (data not shown), suggesting that the negative effect PML/RARα exerts on NIH 3T3 cell growth was due to induction of cell death. The DNA content of the NIH-ErPR2 and the control NIH-Er1 cells was analyzed in the presence and absence of 17βE, by flow cytometry of PI-stained nuclei. The growth curves and representative flow cytometry data of both cell populations are given in Fig. 5 A and B, respectively. The proportion of cycling cells between the NIH-Er and NIH-ErPR cells in either the presence or absence of 17βE demonstrated no marked variations. However, the DNA content of a large proportion (approximately 40%) of nuclei from the NIH-ErPR cells grown in 17βE for 24 and 48 hr was hypodiploid, a feature associated with apoptotic death (14, 17). No correlation between this and the appearance of DNA fragmentation existed, as evaluated by agarose gel electrophoresis (Fig. 5C). The absence of the typical DNA fragmentation pattern was not due to the relatively low percentage of hypodiploid cells in the sample, because the pattern was detected in the DNA extracted from a population of growth factor-deprived TF-1 cells containing a similar proportion of hypodiploid cells (approximately 41%) (Fig. 5C). It therefore would seem that PML/RARα expression in rodent fibroblasts does not cause growth arrest and induces cell death.
Figure 5.
Effects of inducible PML/RARα on survival of NIH 3T3 fibroblasts. NIH-ErPR2 and control NIH-Er1 cells were grown either in the presence or absence of 17βE and analyzed by cell counting (A), flow cytometry (B), and agarose gel electrophoresis (C). For the flow cytometry profiles, cells were stained with PI and evaluated at the indicated time points, in the presence (+) or absence (−) of 17βE. The hypodiploid DNA peak is gated in each profile, and the corresponding percentage of nuclei indicated within the gate is related to the cell death number. This experiment is one of three that gave similar results. For the agarose gel electrophoresis high molecular weight DNA was extracted from NIH-ErPR2 after 48 hr of culture in the presence (+) or absence (−) of 17βE or from TF-1 cells after 48 hr of culture in the presence or absence of granulocyte/macrophage colony-stimulating factor, as indicated.
Cell Death Induction by PML/RARα Depends on the Integrity and Cooperation of the PML Aminoterminal and the RARα Zn Finger Regions.
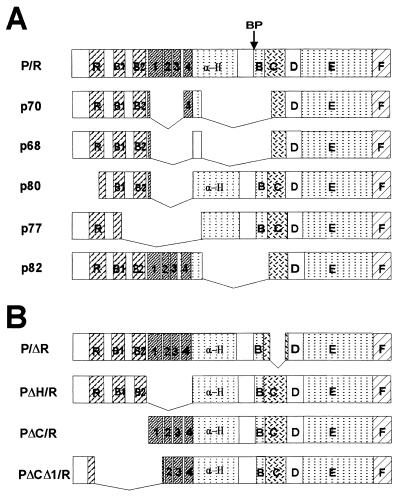
Rare clones obtained from the limiting dilution of PML/RARα-transfected PA317 fibroblasts expressed anti-RARα immunoreactive polypeptides with molecular weights between those of PML/RARα and RARα (not shown). Because they might represent deletion mutants of the PML/RARα protein with impaired capacity to induce cell death, we decided to isolate the corresponding cDNAs from five PA317 cell clones. Nucleotide sequence analysis of the cloned DNAs revealed a single long ORF terminating at the RARα TGA stop codon. Alignment of the various amino acid sequences with that of PML/RARα showed different internal deletions (Fig. 6A). The various PML/RARα mutants can be classified into two groups: (i) mutants with internal deletions involving the PML component of the fusion proteins (p80 and p77), which always affected both the aminoterminal cysteine-histidine-rich region (RING+B1-B2 regions) and the coiled coil region with the four clusters of heptad repeats; and (ii) mutants with internal deletions involving both the RARα Zn finger region and variable portions of the PML α-helix (p82, p70 and p68), including the coiled coil region. Assuming that these mutants are devoid of growth suppressive activity, these results suggest that both PML and RARα sequences contribute to the PML/RARα biological activity. Direct proof for this hypothesis was sought by analyzing the growth inhibitory effect of engineered PML/RARα mutants with deletions in the PML cysteine-histidine-rich region (PΔC/R), the PML coiled coil region (PΔH/R), portions of both the PML cysteine-histidine-rich and coiled coil regions (PΔCΔ1/R), and the RARα Zn finger region (P/ΔR) (Fig. 6B). As shown in Fig. 2C, deletion of the P/ΔR or concomitant deletions of the PΔCΔ1/R abrogated the PML/RARα growth inhibitory activity, whereas individual deletion of the PΔC/R or PΔH/R reduced the biological activity of the fusion protein. These data suggest that the RARα Zn finger and the PML aminoterminal region cooperate toward growth suppression although neither one alone is sufficient for the PML/RARα effect on survival.
Figure 6.
Structural analysis and modular organization of spontaneous and engineered proteins. (A) Schematic representation of the amino acid sequences of PML/RARα and the spontaneous PML/RARα mutants. (B) The indicated PML/RARα mutants were constructed by standard molecular approaches (21). R: RING finger domain. B1, B2: B-boxes. 1, 2, 3, 4: the four clusters of hydrophobic amino acid heptads of the coiled coil region. α-H: α-helix region. BP: PML/RARα junction. B, C, D, E, and F: RARα functional regions. Bridging lines indicates internal deletions.
Cell Death Induction by PML/RARα Depends on its Microspeckled Localization and Matrix Association and Is Not Affected by RA Treatment.
The cellular distribution of PML/RARα is a striking feature of the APL phenotype. RARα is nuclear diffuse, PML is localized within specific subnuclear domains, the so-called PML nuclear bodies (PML-NB), whereas PML/RARα is located in poorly characterized nuclear structures, distinct from the PML-NBs, known as microspeckles. The functional significance of the PML/RARα microspeckles and their relation with subnuclear structures, such as chromatin and matrix, is not known. They are, however, considered critical to the biological activity of the fusion protein, because RA treatment of APL blasts induces terminal differentiation coupled to progressive disappearance of the PML/RARα microspeckles (18–21).
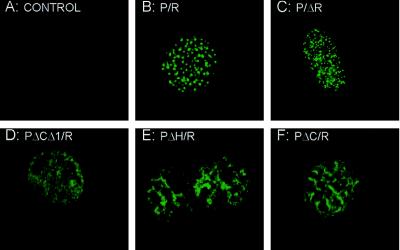
The cellular localization of PML/RARα and the mutated fusion proteins was verified by indirect immunofluorescence using anti-PML (PG-M3 or PG serum, see Materials and Methods) or anti-RARα (RARαF) antibodies and analyzed by confocal laser scanning microscopy. Identical results were obtained in both permissive (stably transfected U937) or nonpermissive (transiently transfected NIH 3T3) cells (representative results of the anti-RARα staining of transfected NIH 3T3 cells are given in Fig. 7). In the absence of the exogenous protein expression, the anti-PML staining of the control NIH 3T3 fibroblasts was negative, because the anti-PML antibodies used in this study did not react with the mouse endogenous PML (Fig. 7A). The PML/RARα and P/ΔR fusion proteins displayed a microspeckled nuclear pattern of localization (Fig. 7 B and C), whereas the PΔCΔ1/R mutant diffusely stained the nucleus (Fig. 7D). The PΔH/R and PΔC/R mutants displayed a nuclear pattern of localization with features that were intermediate between the microspeckled PML/RARα and P/ΔR and the diffuse PΔCΔ1/R fusion protein: fibrous structures and ill-defined areas of homogeneous staining (Fig. 7 E and F).
Figure 7.
Cellular localization of PML/RARα and the various mutant fusion proteins. Immunofluorescence analysis by confocal microscopy of NIH 3T3 cells upon transient transfection of the indicated constructs and staining with the anti-RARα antibody. Control cells (A) were transfected with the empty expression vector.
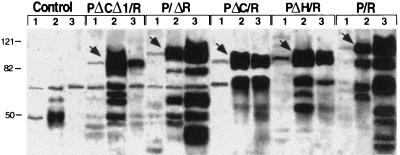
To biochemically characterize the cellular distribution of PML/RARα and the various mutants we analyzed the expression of the fusion proteins by Western blotting in the cytoplasmic, nuclear soluble, and matrix fractions of lysates from cells expressing these fusion proteins. The three fractions were prepared as described in Materials and Methods (16). Comparable results were obtained using NIH 3T3 or U937 cells; results of a typical experiment from stably expressing U937 cells are shown in Fig. 8. Very little PML/RARα or PML/RARα mutant proteins were found in the prenuclear supernatant (cytoplasmic fraction). The PML/RARα and P/ΔR proteins were mainly found in the matrix fraction, whereas the PΔCΔ1/R mutant was mainly detected in the nuclear soluble fraction. The PΔH/R and PΔC/R fusion proteins instead were found both in the nuclear soluble fraction and in the matrix.
Figure 8.
Cell fractionation analysis of PML/RARα and the various mutant fusion protein. Western blot analysis of cellular fractions from lysates of U937 cells stably expressing the indicated fusion proteins. Lanes 1 were loaded with the cytoplasmic fractions, lanes 2 with the nuclear soluble fractions, and lanes 3 with the matrix fractions. The various fusion proteins are indicated by arrowheads. Control cells are U937-transfected with the empty vector (Mt). Blots were stained with the anti-RARα antibody.
It appears that the microspeckled nuclear localization of PML/RARα correlates with the matrix association of the fusion protein and that the integrity of the entire aminoterminal region of PML, including the RING, B1, B2, and coiled coil regions, is required for the localization of the fusion protein to the matrix-associated microspeckles. The nuclear diffuse and soluble PΔCΔ1/R and the microspeckled and matrix-associated P/ΔR mutants are both devoid of any biological activity (Fig. 5), suggesting that localization to matrix-associated microspeckles is necessary, but not sufficient, to the PML/RARα biological activity.
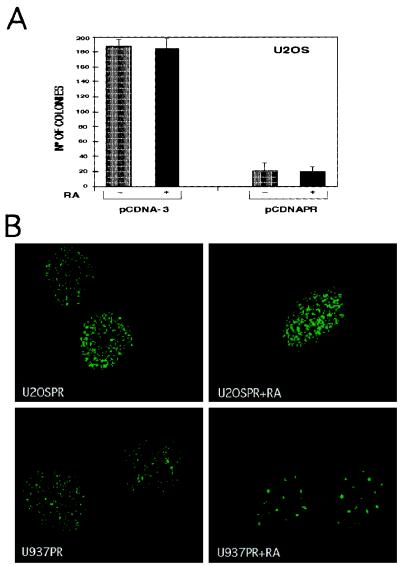
To investigate the effects of RA treatment on the growth suppressive activity of PML/RARα, U2OS cells were transfected with the pCDNAPR (PML/RARa in pcDNA-3) or the empty pCDNA-3 expression vectors, in the presence or absence of RA. After 20 days of G418 selection they were stained with crystal violet, and the number of resistant colonies was counted. RA treatment did not affect the capacity of mock- or PML/RARα-transfected cells to form G418 resistant colonies (Fig. 9A). To determine whether RA treatment induced nuclear redistribution of the fusion protein, we analyzed the localization of PML/RARα in U20S cells by indirect immunofluorescence. To this end, we used a clone of U2OS cells carrying the PML/RARα cDNA under the control of the Zn-inducible MoMt-1 promoter. As a control, the U937MtPR9 clone, in which PML/RARα expression also is induced by Zn treatment, was used. Cells were induced with 100 μM ZnSO4 for 12 hr and then treated with 10−6M RA for 3 days. Cell counting of RA-treated and untreated U2OS cells revealed that RA failed to prevent growth suppression by PML/RARα expression (data not shown). Staining with the anti-PML PGM-3 mAb was performed before and after RA treatment. Both U20S and U937 cells showed the typical microspeckled pattern of PML/RARα localization in the absence of RA treatment (Fig. 9B Left). RA treatment induced the typical speckled relocalization of anti-PML staining into U937 cells, whereas left the PML/RARα microspeckled pattern of U20S cells unchanged (Fig. 9B Right). It therefore appears that RA treatment has no effect on the localization and biological activity of PML/RARα in the nonpermissive U20S cell line.
Figure 9.
Effects of RA treatment on the growth suppressive effect and nuclear localization of PML/RARα. (A) Colony formation assay of U2OS cells in the presence (+) or absence (−) of 10−6 M RA. Cells were transfected either with the pCDNA-3 empty vector or the pCDNAPR vector containing the PML/RARα cDNA. Mean values of an independent triplicate experiment are shown with SD errors. (B) Immunofluorescence analysis of PML/RARα expression in the nonpermissive U2OS (U2OSPR) and in the permissive U937 (U937PR) cell lines. Cells were induced with 100 μM Zn for 12 hr, grown in the presence or absence of RA, and stained with the anti-PML PGM-3 mAb after 3 days of RA treatment.
DISCUSSION
The biological mechanisms through which PML/RARα exerts its leukemogenetic effect in vivo remain unknown. We previously reported two biological activities of PML/RARα into hematopoietic precursor cell lines: block of differentiation and increased survival (6, 8).
In this paper we investigated in greater detail the effects of PML/RARα on cell survival. We found that all the nonhematopoietic cell lines tested were nonpermissive for PML/RARα expression due to a direct growth inhibitory effect of the fusion protein. To investigate the cellular basis of the growth suppression we tried to inducibly express the fusion protein into a variety of nonpermissive cell lines, but succeeded in only two, the NIH 3T3 mouse fibroblast cell line and the U2OS osteosarcoma cell line. In these cells we were able to demonstrate that inducible PML/RARα expression provoked cell death. The inability to inducibly express PML/RARα in most of the nonpermissive cell lines might be the consequence of the modest basal expression leakage encountered in the used inducible expression systems. Notably, the inducible PML/RARα expression obtained in the NIH 3T3 fibroblasts was quite rapidly lost in culture, probably owing to modest PML/RARα basal expression in the absence of the inducer.
The mechanism of cell death (apoptosis or DNA loss after necrotic death) induced by PML/RARα expression into nonpermissive cells remains unclear, because PML/RARα expressing fibroblasts were hypodiploid but did not reveal DNA fragmentation. However, cell death induction appears to be a specific effect of PML/RARα expression into certain cell types because it is not the consequence of cell growth arrest and it was observed with levels of fusion protein expression comparable to those obtained into permissive cells.
Permissivity for PML/RARα expression was variable within the hematopoietic cell types tested: only four (KG-1a, U937, K562, and TF-1) of 10 cell lines tested. We already have reported that PML/RARα increases survival in two of those: it reduces apoptosis in serum-starved U937 and growth factor-deprived TF-1 cells (6, 8). It therefore appears that PML/RARα exerts a cell type specific effect on survival, inducing cell death in nonhematopoietic cell lines and in the majority of hematopoietic cell lines and promoting survival in a subset of hematopoietic cell lines. Overexpression of wild-type PML is associated with suppression of growth (23–26). We have analyzed the effects of PML overexpression in different cell lines and shown that it exerts a growth suppressive effect in all the cell lines used in the present investigation (unpublished work), including those permissive for PML/RARα expression. It is likely that PML/RARα retains the growth suppressor activity of PML, except in certain cell types where the effect of the fusion protein on survival is inverted. The molecular basis for this cell-type specific effect on survival remains however, unclear. We have demonstrated that RA treatment failed to induce the nuclear relocalization of PML/RARα in nonpermissive cell lines, thereby suggesting that the RA signaling pathway of permissive and nonpermissive cells differ.
The cell type-specific effect PML/RARα exerts on cell survival may account for the specificity of the t(15;17). This translocation is unique to APL and has never been reported in other neoplasms (reviewed in ref. 8). The molecular basis of the tissue specificity of the t(15;17) is not known. The chromosome translocations associated with lymphoid neoplasms invariably involve the Ig or T cell receptor loci, and their lineage specificity is thought to be the consequence of the genetic rearrangements that regulate Ig or T cell receptor gene expression in lymphoid tissues (22). PML and RARα are widely expressed genes, but they are not known to exert hematopoietic specific functions. It could be that the t(15;17) is equally likely to occur in cells from different tissues, but that it is selected by the neoplastic phenotype only when it occurs in permissive hematopoietic cells. The fact that PML/RARα permissivity is restricted to a subset of hematopoietic cell lines suggests that it varies even within the hematopoietic compartment. Notably, not all hematopoietic-specific promoters used in the attempts to generate PML/RARα transgenic mice have permitted a leukemic phenotype to be expressed (A. Dejean, personal communication; refs. 4 and 5), suggesting that targeting of the PML/RARα protein to a specific compartment of the myeloid stem cells is a prerequisite for its leukemogenetic effect.
The capacity of PML/RARα to induce cell death in nonpermissive cell lines appears to depend on both PML and RARα sequences. Independent deletions of the RARα Zn finger and the PML aminoterminal regions completely abrogated the growth inhibitory effect of the fusion protein. Individual deletions of the PML aminoterminal region including the RING, B1 and B2 domains, or the coiled coil region only partially affected the biological activity of PML/RARα. We previously reported that the integrity of the PML coiled coil region is absolutely required for the capacity of PML/RARα to block differentiation in vitro, whereas the RARα Zn finger is only partially required and the PML RING and B1 and B2 domains are dispensable (13). It therefore appears that different molecular mechanisms regulate the two biological activities of PML/RARα, e.g., block of differentiation and growth suppression.
The mechanisms by which the deletions of the RARα Zn finger and the PML aminoterminal regions abrogate the capacity of PML/RARα to induce cell death remain unknown. The RARα Zn finger is a sequence-specific DNA binding domain through which PML/RARα regulates RA target genes (1, 11, 12). Consequently, the capacity of PML/RARα to induce cell death appears to involve regulation of RA target genes. The RA target genes that are regulated by PML/RARα have not yet been identified. Interestingly, we recently have reported that type II transglutaminase, a protein involved in the regulation of growth and apoptosis, is directly regulated by PML/RARα (27). The function of the RING finger region and the B1 and B2 boxes is unknown. Their integrity is required for PML-nuclear body formation, and three-dimensional structures suggest that they are interfaces for interaction with other proteins (9, 10, 26). The PML coiled coil region is a protein–protein interaction domain that, in wild-type PML, mediates dimerization both with itself and PML/RARα (11). We have demonstrated that the microspeckled PML/RARα fusion protein mainly is associated with the nuclear matrix and that this association depends on the integrity and cooperation of the PML RING+B1-B2 and coiled coil regions, suggesting that the PML aminoterminal region is crucial for the targeting of the fusion protein to specific, matrix-associated nuclear subdomains, the microspeckles, possibly through multiple protein–protein interactions. Because the deletion of the PML aminoterminal region of the fusion protein provoked loss of microspeckled localization, matrix association and cell death induction effect, it appears that the microspeckles are the active sites of the PML/RARα activity on survival. Targeting of the PML/RARα fusion protein to the matrix-associated microspeckles, however, does not appear to be sufficient for the capacity of PML/RARα to induce cell death, because deletion of the RARα Zn finger region impaired the PML/RARα biological activity without interfering with its localization. In conclusion, our results suggest that the targeting of RARα sequences to matrix-associated microspeckles by PML sequences is crucial for the capacity of the fusion protein to induce cell death of certain cell types.
Acknowledgments
We thank T. Casini and C. Calvio for helpful discussion, P. P. Di Fiore for critical reading of the manuscript, and D. Riganelli for support in the computer acquisition and analysis of immunofluorescence images. P.F.F. is supported by a Fondetione Istituto Europeo di Oncologia fellowship. This work has been supported by grants from Associazione Italiana per la Ricerca sul Cancro, Consiglio Nazionale delle Ricerche–Applicazioni Cliniche della Ricerche Oncologica, and European Community (Biomed and Biotech).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: PML, promyelocytic leukemia; APL, acute promyelocytic leukemia; RARα, retinoic acid receptor; RA, retinoic acid; PI, propidium iodide; PΔC/R, PML cysteine-histidine-rich region; PΔH/R, PML coiled coil region; PΔCΔ1/R, both the PML cysteine-histidine-rich and coiled coil regions; P/ΔR, RARα Zn finger region.
References
- 1.Grignani Fr, Fagioli M, Alcalay M, Longo L, Pandolfi P P, Donti E, Biondi A, Lo Coco F, Grignani F, Pelicci P G. Blood. 1994;83:10–15. [PubMed] [Google Scholar]
- 2.Warrel R P, Jr, de Thé H, Wang Z Y, Degos L. New Engl J Med. 1993;329:177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 3.Altabef M, Garcia M, Lavau C, Bae S, Dejean A, Samaruth J. EMBO J. 1996;15:2707–2716. [PMC free article] [PubMed] [Google Scholar]
- 4.Grisolano J L, Wesselschmidt R L, Pelicci P G, Ley T J. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 5.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grignani Fr, Ferrucci P F, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C, Nicoletti I, Pelicci P G. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 7.Grignani Fr, Testa U, Fagioli M, Barberi T, Masciulli R, Mariani G, Peschle C, Pelicci P G. Cancer Res. 1995;85:440–443. [PubMed] [Google Scholar]
- 8.Rogaia D, Grignani F, Grignani F, Nicoletti I, Pelicci P G. Leukemia. 1995;9:1467–1472. [PubMed] [Google Scholar]
- 9.Borden K L, Boddy M N, Lally J, O’Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borden K L, Lally J M, Martin S R, O’Reilly N J, Solomon E, Freemont P S. Proc Natl Acad Sci USA. 1996;93:1601–1606. doi: 10.1073/pnas.93.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. EMBO J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nervi C, Poindexter C E, Grignani F, Pandolfi P P, Lo Coco F, Avvisati G, Pelicci P G, Jetten A M. Cancer Res. 1992;52:3687–3692. [PubMed] [Google Scholar]
- 13.Grignani Fr, Testa U, Rogaia D, Ferrucci P F, Samoggia P, Pinto A, Aldinucci D, Gelmetti V, Fagioli M, Alcalay M, Seeler J, Grignani F, Nicoletti I, Peschle C, Pelicci P G. EMBO J. 1996;15:101–110. [PMC free article] [PubMed] [Google Scholar]
- 14.Nicoletti I, Migliorati G, Pagliacci M C, Grignani F, Riccardi C. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 15.Flenghi L, Fagioli M, Tomassoni L, Pileri S, Gambacorta M, Pacini R, Grignani Fr, Casini T, Ferrucci P F, Martelli M F, Pelicci P G, Falini B. Blood. 1995;85:1871–1880. [PubMed] [Google Scholar]
- 16.Lee K A W, Green M. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- 17.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz M O, Lassota P, Traganos F. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 18.Daniel M T, Koken M, Romagne O, Barbey S, Bazarbachi A, Stadler M, Guillemin M C, Degos L, Chomienne C, de Thé H. Blood. 1993;82:1858–1867. [PubMed] [Google Scholar]
- 19.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Koken M H M, Puvion-Dutilleul F, Guillemin M C, Viron A, Cruz-Linares G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de Thè H. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis K, Rambaud S, Lavau C, Jansen J, Carcalho T, Carmo-Fonseca M, Lamond A, Dejean A. Cell. 1994;76:345–358. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 22.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 23.Koken M H M, Cruz-Linares G, Quignon F, Viron A, Chelbi-Alix M K, Sobczak-Thèpot J, Juhlin L, Degos L, Calvo F, de Thèf H. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 24.Liu J H, Mu Z M, Chang K S. J Exp Med. 1995;1815:1965–1973. doi: 10.1084/jem.181.6.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu Z M, Chin K V, Liu J H, Lozano G, Chang K S. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le X F, Yang P, Chang K S. J Biol Chem. 1996;271:130–135. doi: 10.1074/jbc.271.1.130. [DOI] [PubMed] [Google Scholar]
- 27.Benedetti L, Grignani F, Scicchitano B M, Jetten A M, Diverio D, Lo Coco F, Avvisati G, Gambacorti-Passerini C, Adamo S, Levin A A, Pelicci P G, Nervi C. Blood. 1996;87:1939–1950. [PubMed] [Google Scholar]