Abstract
Inorganic arsenic, a human carcinogen, is enzymatically methylated for detoxication, consuming S-adenosyl-methionine (SAM) in the process. The fact that DNA methyltransferases (MeTases) require this same methyl donor suggests a role for methylation in arsenic carcinogenesis. Here we test the hypothesis that arsenic-induced initiation results from DNA hypomethylation caused by continuous methyl depletion. The hypothesis was tested by first inducing transformation in a rat liver epithelial cell line by chronic exposure to low levels of arsenic, as confirmed by the development of highly aggressive, malignant tumors after inoculation of cells into Nude mice. Global DNA hypomethylation occurred concurrently with malignant transformation and in the presence of depressed levels of S-adenosyl-methionine. Arsenic-induced DNA hypomethylation was a function of dose and exposure duration, and remained constant even after withdrawal of arsenic. Hyperexpressibility of the MT gene, a gene for which expression is clearly controlled by DNA methylation, was also detected in transformed cells. Acute arsenic or arsenic at nontransforming levels did not induce global hypomethylation of DNA. Whereas transcription of DNA MeTase was elevated, the MeTase enzymatic activity was reduced with arsenic transformation. Taken together, these results indicate arsenic can act as a carcinogen by inducing DNA hypomethylation, which in turn facilitates aberrant gene expression, and they constitute a tenable theory of mechanism in arsenic carcinogenesis.
Arsenic is a human carcinogen and the adverse effects from exposure to this metalloid are considered among the top priority hazards in the United States. In exposed populations, arsenic is associated with various tumors, including tumors of the lung, skin, bladder, and liver (1, 2). However, arsenic as a carcinogen remains an enigma because, on the one hand, it is definitively active in humans, whereas on the other, carcinogenesis in rodent models has never been convincingly demonstrated. Consequently, it appears that humans are particularly sensitive to arsenic-induced malignancies, at least when compared with rodent species. The lack of knowledge of the carcinogenic mechanism of action together with the apparent sensitivity of human populations creates even more concern for the adverse potential of this important environmental pollutant.
Arsenic as an inorganic compound has a distinctive metabolism in mammals in that it undergoes enzymatic mono- and dimethylation in what is generally considered to be a detoxication pathway (3, 4). Inorganic arsenic exists in both a pentavalent form, arsenate, and a trivalent form, arsenite, the latter being more toxic. Both valence forms of arsenic undergo enzymatic methylation. Arsenic methylation requires S-adenosyl-methionine (SAM) as a cofactor and, as yet, largely uncharacterized methyltransferases (MeTases). Humans are effective methylators of arsenic and methylation occurs at high levels in the liver, a suspected target organ for arsenic carcinogenesis (1, 2). SAM, as a methyl group donor, is an essential cofactor for a variety of MeTases, including DNA MeTases, a group of enzymes responsible for DNA methylation. DNA methylation status contributes to the control of the expression of a variety of genes, including several oncogenes (5–8). DNA hypomethylation is thought to constitute an early event in some cancers (7) and has been associated with many types of tumor (5, 6, 9, 10).
This work tested the hypothesis that, because arsenic biotransformation consumes cellular methyl groups, chronic arsenic exposure could induce DNA hypomethylation, causing aberrant gene expression to occur which, in turn, would facilitate transformation. This would constitute an epigenetic mechanism for arsenic carcinogenesis that would be consistent with its poor mutagenicity (11–14).
MATERIALS AND METHODS
Cell Line and Sodium Arsenite Exposure.
The cell line used (TRL 1215) was originally derived from the liver of 10-day old Fischer F344 rats (15). The cells are diploid and normally nontumorigenic. Cells were cultured with passage once per week in Williams’ medium E containing 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine with the addition of the following amounts of sodium arsenite (0, 0.125, 0.250, and 0.500 μM). The medium was changed at 3-day intervals. Sodium arsenite stock solution was prepared with deionized distilled water and sterile filtered. The final concentrations of arsenite were obtained by the appropriate dilution with media. Cells were visually monitored for signs of morphological transformation at weekly intervals. Initial experiments confirmed the capacity of TRL 1215 cells to methylate arsenic.
Metabolic Integrity Assay.
A Promega Cell Titer 96 Non-Radioactive Cell Proliferation/Cytotoxicity Assay kit was used to quantify the cytotoxicity of sodium arsenite in TRL 1215 cells by assessment of metabolic integrity. The assay measures the amount of a formazan produced by metabolic conversion of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide by dehydrogenase enzymes located in the intact mitochondria of viable cells.
Tumor Growth in Athymic Nude Mice.
Cells treated for 8 weeks with 0 or 0.500 μM or for 18 weeks with 0, 0.125, 0.250, and 0.500 μM sodium arsenite were trypsinized and collected in 0.9% saline at a concentration of 2 × 106 cells per 200 μl. Athymic Nude (NCr-nu) mice (National Cancer Institute–Frederick Cancer Research and Development Center Animal Production Area, Frederick, MD) were grouped (n = 13–15) according to in vitro arsenic dosage and inoculated subcutaneously in the dorsal thoracic midline with 2 × 106 cells. The development of tumors was accessed over the next 5 months by palpation and confirmed by light microscopy after necropsy. Rate of metastasis was determined by visual inspection of the lungs at necropsy.
Construction of Rat Satellite I DNA Probe.
A pCR-J00784 plasmid containing a 235-bp DNA fragment from rat satellite I DNA GB-RO:J00784, which contains two copies of the 92–93 bp repetitive fragments was constructed in our laboratory. The PCR product of this DNA fragment was first synthesized by using normal, untransformed TRL 1215 cell DNA as the template. Primers for the PCR amplification of the repetitive fragment were chosen through genebank (Genetics Computer Group, Madison, WI). The PCR product was subsequently cloned into pCR II plasmid by using Original TA-cloning kit (Invitrogen). The satellite repeats, contained in a BamHI–XhoI digested fragment, were electrophoretically separated from the plasmid DNA on a 2% NuSieve GTG gel (FMC). The BamHI/XhoI band was then excised and the DNA was electroeluted and ethanol purified.
DNA Methylation and Southern Blot Analysis.
Genomic DNA was isolated and purified according to the method of Laird et al. (16) with slight modification. Purified genomic DNA (10 μg) was digested by HpaII and MspI by using 4 units of enzyme per μg of DNA to completion at 18 hr. DNA was then precipitated by ethanol, redissolved in H2O, separated electrophoretically with 0.7% SeaKem Agarose gel (FMC) and blot transferred to Maximum Strength NYTRAN membrane (Schleicher & Schuell) after depurination. The DNA was cross-linked by UV irradiation and the membrane was baked for 2 hr at 80°C. Hybridization was accomplished at 65°C for 15–18 hr in hybridization solution containing the pCR-J00784 probe labeled with [32]dCTP (Amersham) using a Multiprime DNA random-labeling system kit (Amersham). The membranes were washed twice at high stringency for 15 min at room temperature with 2× SSC/0.1% SDS and three times at 65°C for 60 min with 0.1× SSC/0.1% SDS. Bands were then visualized autoradiographically and analyzed densitometrically with ImageMaster VDS version 2.0 (Pharmacia Biotech).
Isolation and Northern Blot Analysis of RNA.
Total RNA was isolated by using the RNAzol method (Tel-Test, Friendswood, TX). Aliquots of RNA were assessed for integrity by electrophoresis and ethidium bromide staining for visualization of the 28S and 18S rRNA bands. Northern blot analysis of the mRNA was performed as described (17). The resulting Northern blots were hybridized with 32P random-labeled mouse DNA MeTase fragment for DNA MeTase mRNA. The mouse DNA MeTase Gb-Ro:X14805 fragment (4351–4553), which is considered to be the conserved region for the catalytic activity (18), was used for assessing the level of DNA MeTase mRNA. T7 RNA polymerase was used for riboprobe labeling with ribonuclear labeling kit for the transcription of the antisense RNA probe of mouse DNA MeTase and 18S rRNA (template from Ambion, Austin, TX). 18S rRNA was used to standardize total RNA loading.
Metallothionein (MT) Protein Assay.
Cells were grown in normal medium to 50% confluence. Media containing different concentrations of cadmium, a known inducer of MT expression (19), were then added. After 24 hr, cells were harvested by trypsinization, counted, and ruptured by sonication. MT protein levels were then estimated using the Cd saturation assay (20). Values were normalized to cell numbers and the control value was set at 100%. In spontaneously transformed TRL 1215 cells, MT protein levels were measured without previous exposure to inducing stimulus.
DNA MeTase Enzyme Activity Assay.
A modified assay based on the work of Adams et al (21) was used to determine DNA MeTase activity. Cell lysates containing 50 μg of total protein were incubated at 37°C for 1 hr with a deoxyinosine–deoxycytosine double-stranded DNA template (poly[dI⋅dC]⋅poly[dI⋅dC]; Sigma) and 3H-labeled S-adenosyl-methionine (SAM) (Amersham). Reactions were stopped by the addition of 1% SDS and 150 μg proteinase K for 2 hr at 60°C. The DNA template was recovered by trichloroacetic acid (TCA) coprecipitation with 125 μg sheared salmon sperm DNA. RNA was removed by resuspension of the precipitates in 0.5 M NaOH. DNA template was precipitated and washed with 10% TCA. After drying, the DNA pellet was placed in scintillation mixture and activity was determined by liquid scintillation counting. The DNA MeTase activity was analyzed as dpm/μg of total protein and then normalized to control. All assays were performed in triplicate.
Quantitative Analysis of SAM and SAH.
An HPLC system (Hewlett–Packard 1090), coupled with C-18 ODS ion pairing reverse-phase column (Advantage-60, 5 μm, 60 Å, 4.6 × 250 mm; Thomson, Chantilly, VA) was used for the separation and quantitation of SAM and SAH. S-adenosyl-ethionine was used as internal standard. A gradient elution system was used with modified linear gradient for the separation of SAM, SAH, and S-adenosyl-ethionine (22).
RESULTS
The initial goal of these studies was to develop a tenable cell model to study the molecular events occurring during arsenic carcinogenesis. To this end, TRL 1215 cells were chronically exposed to subtoxic level of sodium arsenite. Low, subtoxic levels duplicate human exposure situations and avoid extraordinary, nonphysiological responses potentially induced by doses of high toxicity.
Cytotoxicity of Sodium Arsenite in TRL 1215 Cells.
To establish the appropriate level of exposure for chronic transformation by arsenic, the acute cytotoxic effects of arsenic were defined in TRL 1215 cells by assessment of metabolic integrity. Arsenic showed a typical sigmoidal increase in toxicity verses concentration, and the LC50 was determined to be 3.43 μM sodium arsenite. In subsequent transformation studies, to eliminate any possible overt cytotoxic effects, 0.125, 0.250, and 0.500 μM concentrations of sodium arsenite were used. The highest dose selected is less than 1/7 of the LC50.
Malignant Transformation of TRL 1215 Cells by Sodium Arsenite.
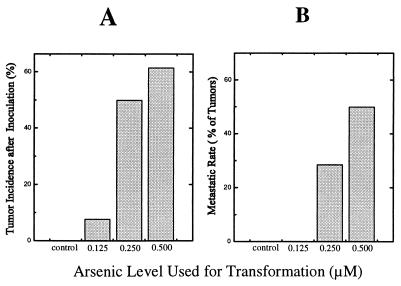
Cells grown in the presence of the highest concentration of sodium arsenite (0.500 μM) started exhibiting morphological changes indicative of transformation as early as the eighth week of exposure. A morphological change of the cells from epithelioid to fibroblast-like occurred in a small portion (≈5%) of the cells at this point, whereas unexposed cells showed no such changes (not shown). However, when cells treated with 0.500 μM arsenic for 8 weeks were collected and inoculated into Nude mice no tumors developed (0 tumors per 15 mice inoculated), in a fashion similar to unexposed cells (0/15), clearly showing that malignant transformation had not yet occurred. In subsequent weeks, the cellular morphological changes showed graded increases. After 18 weeks of continuous exposure to arsenic, when morphological changes had become much more frequent (≈70% of cells at the highest dose), inoculation of cells into Nude mice gave rise to tumors in an arsenic dose-dependent manner (Fig. 1A). These results indicate that malignant transformation of the TRL 1215 cells with arsenic had occurred by 18 weeks of exposure. Assuming a linear dose relationship for arsenic and malignant transformation between these two established time points, the exposure duration for which significant (P ≤ 0.05 by Fisher exact test, n = 15) increases in malignant transformation (33% tumor incidence after inoculation) had first occurred can be estimated at 13.4 weeks. Tumors resulting from inoculation of TRL 1215 cells malignantly transformed by arsenic showed both fibrosarcomatous and undifferentiated areas with multiple mitotic features and frequent invasion into the subdermal muscle layers. These tumors also showed a high proportion of metastases to the lung (Fig. 1B). Additionally, when cells malignantly transformed by 18 weeks of exposure to arsenic were cultured in the absence of arsenic for 6 weeks, the cell morphology remained distinctly different from control and, when inoculated into Nude mice, these cells also gave rise to aggressive, malignant tumors.
Figure 1.
Tumorigenicity of arsenic-exposed TRL 1215 cells in nude mice. TRL 1215 cells treated with 0, 0.125, 0.250, and 0.500 μM arsenic for 18 weeks were inoculated subcutaneously into Nude mice (n = 13–15). (A) Tumor incidence data as a percentage of the number of mice inoculated. (B) Rate of metastasis as a percentage of total tumors.
Arsenic Transformation and DNA Hypomethylation.
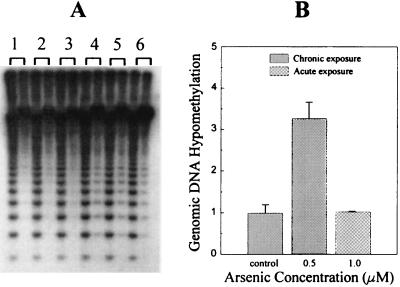
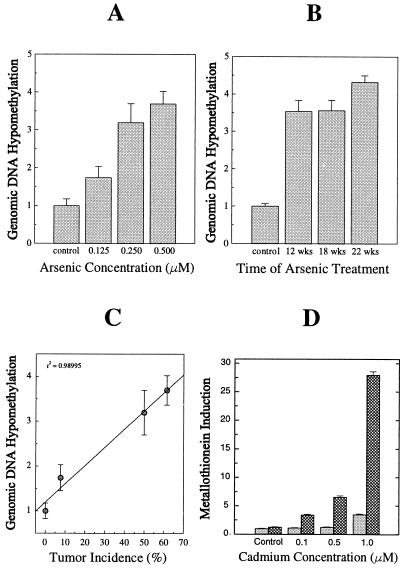
Using the methylation-sensitive restriction endonuclease isoschizomers, HpaII and MspI, global DNA hypomethylation was assessed. Initially, genomic DNA from TRL 1215 cells treated with a low dose of arsenite (0.5 μM) for 18 weeks and with higher doses of arsenite (1.0 μM) acutely for 48 hr were compared. DNA from arsenic-transformed cells was much more easily digested by methylation-sensitive restriction endonuclease HpaII than DNA from untransformed cells (Fig. 2A). The extent of DNA hypomethylation was densitometrically analyzed and compared using MspI cutting bands to standardize DNA loading. HpaII digestion ability increased at least 3-fold for DNA from arsenic-transformed cells. The hypomethylation of genomic DNA was specifically related to the chronic treatment with arsenic, because no change in methyl sensitive restriction enzyme digestion was observed in acutely exposed cells (Fig. 2B). The increase in genomic DNA hypomethylation was dependent on arsenic dose and duration of exposure (Fig. 3 A and B). The dose-dependent increases in DNA hypomethylation showed a highly significant (P = 0.01) positive correlation with the dose-dependent increases in malignant transformation as assessed by tumor formation after inoculation (Fig. 3C), further implicating the association between these two events. Genomic DNA methylation status in transformed cells, which were cultured in arsenic-free medium for an additional 6 weeks, showed that DNA remained similarly hypomethylated in the same dose-dependent fashion. This indicates that an altered methylation pattern persists even in the absence of arsenic and is consistent with the production of tumors in Nude mice from the inoculation of cells grown for a period in arsenic-free media after arsenic transformation had occurred.
Figure 2.
Arsenic-induced alterations in global DNA methylation. Global DNA methylation as determined in genomic DNA isolated from control and arsenic-transformed cells. (A) Isolated DNA digested with MspI (left lane of each numbered group) and its methyl-sensitive isoschizomer HpaII (right lane of each numbered group), followed by Southern blot analysis using rat satellite I DNA repeat fragment probe pCR-J00784. Groups: 1, control cells at 18 weeks; 2 and 3, duplicates of cells acutely treated with 1.0 μM arsenic; 4–6, triplicates of arsenic-transformed cells with 0.5 μM arsenic for 18 weeks. (B) Densitometric analysis of the results of the Southern blots from control and chronic (18 weeks) or acute (48 hr) arsenic exposure at the concentrations indicated. Data represent mean ± SEM of triplicate experiments.
Figure 3.
Dose- and exposure duration-dependence of arsenic-induced global DNA hypomethylation and altered gene expression in arsenic-transformed cells. (A) DNA methylation status in cells treated with arsenic for 19 weeks and cultured in arsenic-free medium for 6 more weeks. (B) DNA methylation status in cells exposed to 0.5 μM arsenic at times indicated. All data are normalized to the control. (C) Correlation based on the various dosage groups of arsenic between genomic DNA methylation status and tumor incidence after inoculation. (D) MT expression in arsenic-transformed cells (cross-hatched column) and nontransformed cells (solid column) determined after cells were exposed to cadmium at the indicated concentration for 24 hr. Data are normalized to nontransformed cells without exposure to cadmium and represent mean ± SEM of triplicate experiments.
Consistent with DNA hypomethylation, chronic arsenic exposure caused a significant (P < 0.05) depletion (19.3 ± 6.2%) of SAM in arsenic-transformed cells when compared with nontransformed cells (SAM level 74.2 ± 7.8 pmol/106 cells). Levels of the metabolic by-product of MeTase utilization of SAM as a cofactor, SAH, were unchanged (transformed 18.0 ± 1.8 pmol/106 cells; untransformed 18.7 ± 1.2 pmol/106 cells). However, a significant (P < 0.05) reduction (19.9 ± 5.9%) in the SAM/SAH ratio in arsenic-transformed cells did occur.
MT Hyperinducibility in Arsenic-Transformed Cells.
The expressibility of the MT gene is clearly dependent on the DNA methylation status (8). Although DNA hypomethylation alone is able to modestly elevate MT expression, MT inducers such as cadmium will cause a marked hyperinducibility when the MT gene is hypomethylated by various means, including treatment with 5-aza-cytidine (8, 23). In arsenic-transformed cells, basal levels of MT showed a significant increase over control levels (27.7 ± 3.4%). Additionally, in arsenic-transformed cells, MT levels were induced up to 22-fold above basal levels when exposed to an inducing stimulus (1.0 μM cadmium), whereas in nontransformed cells the same stimulus only produced a 3.5-fold increase (Fig. 3D). Thus, DNA hypomethylation in arsenic-transformed cells enhanced basal expression and markedly increased inducibility of MT protein production. In contrast, in a TRL 1215 subpopulation of cells that had undergone spontaneous transformation subsequent to repeated passages (in excess of 24), the constitutive expression of the MT gene was highly depressed (8.31 ± 4.07 μg MT protein per 106 cells) compared with untransformed cells (61.0 ± 18.1 μg MT protein per 106 cells). Thus, spontaneously transformed TRL 1215 cells showed clear phenotypic differences from arsenic-transformed TRL 1215 cells.
DNA MeTase Activity Is Reduced in Arsenic-Transformed Cells.
DNA MeTase is involved in both maintenance of methylation status and restoration of demethylated sites. It has also been reported that overexpression of DNA MeTase is associated with certain tumor cell lines and/or early events in cancer (24). DNA MeTase enzyme activity in arsenic-transformed cells was depressed by up to 40% after transformation by arsenic (Table 1). DNA MeTase activity was not significantly reduced by acute arsenic exposure. The steady-state expression of DNA MeTase gene was surprisingly increased by up to 2-fold after arsenic transformation. These results suggest that chronic exposure to arsenic results in a dose-dependent loss of DNA MeTase activity, possible due to a decrease in SAM/SAH ratio, and in an attempt to compensate for the decrease in activity, expression of the DNA MeTase gene was elevated.
Table 1.
DNA MeTase enzyme and gene activity
| Arsenic concentration, μM | DNA MeTase activity
|
DNA MeTase mRNA level (18 weeks) | |
|---|---|---|---|
| Chronic exposure (18 weeks) | Acute exposure (24 hr) | ||
| 0 | 100 ± 11.9 | — | 100 |
| 0.125 | 95.3 ± 12.5 | — | 145 |
| 0.250 | 79.0 ± 7.4 | — | 164 |
| 0.500 | 59.7 ± 6.0* | 78 ± 4.5 | 212 |
All data are normalized to the control value and are given as a percentage. Asterisk indicates a value significantly different (P ≤ 0.05) from control.
DISCUSSION
Arsenic, as an environmental agent, is considered to be a very high priority toxic substance largely due to its carcinogenic potential in humans (1, 2). The perplexity presented by the clear human carcinogenic capacity of this metalloid in the absence of substantiating rodent data creates further concern over what might be distinctive sensitivity in humans. In this case, defining potential mechanisms is critical in defining the nature and extent of the human health hazard. This study demonstrates that epithelial cells will undergo malignant transformation after chronic, low-level arsenic exposure and that this transformation is associated with DNA hypomethylation and aberrant gene expression. The latter includes the increased basal level expression and hyperexpressibility upon stimulation of the MT gene. The expression of the MT gene has been definitively linked to methylation status (8, 23). The association of DNA hypomethylation with carcinogenesis has been repeatedly demonstrated with other chemical agents or with methyl-restricted diets (9, 25) and was predicted based on our hypothesis that arsenic would consume cofactors essential for duplication and maintenance of DNA methylation status. These results provide the foundation for a tenable theory for mechanism in arsenic carcinogenesis and are consistent with data indicating arsenic is largely ineffective as a mutagen (11–14). In light of these findings associating arsenic methylation and malignant transformation, the fact that humans are effective methylators of arsenic may play a critical role in defining sensitivity to its carcinogenesis. The methylation of arsenic shows distinct species differences. In humans exposed to arsenic, nearly 90% of the total urinary arsenic has undergone methylation before excretion. This sharply contrasts with a level of only 20% of total urinary arsenic appearing as methylated species in rat urine (26), whereas in mice methylation of arsenic is also substantially lower than in humans (27). The TRL 1215 cells used for transformation are able to methylate arsenic at levels more typical for the rat. The consumption of methyl groups in arsenic biotransformation presumably would affect DNA methylation in a fashion related to the extent of arsenic methylation, thus potentially accounting for species differences in carcinogenic sensitivity.
Global hypomethylation of DNA has been demonstrated in various human tumors (7, 10) and during rat or mouse hepatocarcinogenesis (9, 25). DNA hypomethylation is, in fact, thought to be an early event in human carcinogenesis (7) and is associated with genetic instability in cancer cells (28). Similarly, changes in DNA methylation patterns are common in cultured tumor cell lines and primary tumor cells (5, 6). Our results show that genomic DNA methylation was substantially decreased concurrently with arsenic transformation, and that these decreases are dose related, dependent on exposure intervals, and inheritable. All these factors are consistent with DNA methylation as an epigenetic mechanism for carcinogenesis in which aberrant gene expression could alter phenotype in the absence of gene mutation (25, 29–31). Alterations in DNA methylation patterns can be fixed during DNA replication and evidence suggests that methylation status of a gene can participate, with other gene control mechanisms, to balance the transcriptional level of the gene (25). Hypomethylation is associated with activation of various oncogenes during cancer development, such as c-myc, c-fos, and H-ras in human and rat liver tumors (5, 6) and raf (32) in mouse liver tumors. Additionally, c-myc overexpression is correlated with its specific hypomethylation in HL-60 cells (33) and in early rat liver tumors (34). Chronic depletion of cellular SAM contents or blockade of SAH hydrolysis, which both result in reductions of SAM/SAH ratio, also result in an overexpression of c-myc (33, 34). DNA hypomethylation occurring during hepatocarcinogenesis with nongenotoxic agents can similarly facilitate oncogene activation (25). Although acute arsenic exposure induces expression of a variety of genes (35–39), including oncogenes, it is clear that such activations are not due to the direct DNA effects such as base mutation. Arsenic, at nontoxic doses, is typically nongenotoxic and evidence indicates the metalloid generally does not cause mutations in either mammalian or bacterial systems (11–14). The evidence for the existence of a threshold for arsenic carcinogenesis in human populations (40, 41) is similarly in keeping with an epigenetic mechanism of carcinogenesis.
The expression of MT genes is clearly dependent on DNA methylation status (8) and the gene activity and responsiveness to inducing stimuli are dramatically increased after exposure to hypomethylating agents, such as the pyrimidine analogue 5-aza-cytidine (8, 21, 40). If the 5′-flanking regions of the MT gene are highly methylated, it will be poorly expressed and minimally reactive to inducing stimuli, making the inducibility of this gene a strong indicator of DNA hypomethylation, as recently demonstrated (42, 43). Our results clearly indicate that in arsenic-transformed cells, the MT gene is hyperinducible, indicative of hypomethylation as a basis for aberrant gene expression. In sharp contrast, the MT gene is actually down-regulated in liver tumors induced with the highly mutagenic, alkylating carcinogen, N-nitrosodiethylamine (44, 45), or in TRL 1215 cells that have undergone spontaneous transformation with repeated passage, an event likely due to spontaneous mutation and subsequent clonal expansion (46). This indicates that the hyperinducibility of the MT gene seen in association with arsenic-induced transformation is not simply due to DNA hypomethylation occurring during rapid cell proliferation from limitations in the capacity or fidelity of DNA maintenance methylation (47). Taken together, these results strongly point toward hypomethylation of DNA as the causative factor in arsenic-induced malignant transformation.
The reduced DNA MeTase activity that also occurred with arsenic transformation is intriguing, particularly in concert with the enhanced transcription of the DNA MeTase gene. As a general finding, arsenic can be an effective enzyme inhibitor, and can inhibit a variety of enzymatic processes (48). Alternatively, with continuous exposure to a methylation substrate such as arsenic, the cell could produce greater quantities of SAH, the byproduct of methylations using SAM as the methyl donor. SAH is an effective competitive product inhibitor of DNA MeTase activity (49, 50). Steady-state levels of SAH were not, however, increased in arsenic-transformed cells although information on both rates of production and of consumption of SAH would be required to make definitive statements about any role in decreasing DNA MeTase activity. However, the SAM/SAH ratio, which appears to be an important regulator of transmethylation reactions using SAM, was substantially decreased in arsenic-transformed cells. A decreased SAM/SAH ratio may produce an inhibition of most of the important MeTases, such as DNA MeTase (51). Whatever the precise causes, it appears that multiple factors involving the arsenic metabolism, both directly and indirectly, may contribute to the final level of DNA methylation in arsenic-induced malignant transformation.
Acknowledgments
We wish to thank Drs. Marianno Cebrián, Betzabet Quintanilla-Vega, and Luz Maria Del Razo for their assistance on the measurement of metabolites of arsenic; Jim Hochadel, Robert Bare, and George Smith for technical assistance; and Drs. Yih-horng Shiao, R. Rita Misra, Gregory S. Buzard, and T. Phillip Waalkes for their valuable suggestions.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: MeTase, methyltransferase; MT, metallothionein; SAH, S-adenosyl-homocysteine; SAM, S-adenosyl-methionine.
References
- 1.International Agency for Research on Cancer. IARC Monograph on the Evaluation of Carcinogenic Risks to Humans–Overall Evaluations of Carcinogenicity: An Update of IARC Monographs 1 to 42. Lyon, France: IARC; 1987. , Suppl. 7, pp. 100–106. [Google Scholar]
- 2.Waalkes M P. In: Metal Toxicology. Goyer R A, Klaassen C D, Waalkes M P, editors. New York: Academic; 1995. pp. 54–56. [Google Scholar]
- 3.Zakharyan R A, Wildfang E, Aposhian H V. Toxicol Appl Pharmacol. 1996;140:77–84. [PubMed] [Google Scholar]
- 4.Marafante E, Vahter M. Environ Res. 1987;42:72–82. doi: 10.1016/s0013-9351(87)80008-7. [DOI] [PubMed] [Google Scholar]
- 5.Rao P M, Antony A, Rajalakshmi S, Sarma D S R. Carcinogenesis. 1989;10(5):933–937. doi: 10.1093/carcin/10.5.933. [DOI] [PubMed] [Google Scholar]
- 6.Nambu S, Inoue K, Sasaki H. Jpn J Cancer Res (Gann) 1987;78:695–704. [PubMed] [Google Scholar]
- 7.Vogelstein B, Fearon E R, Hamilton S R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M M, Bos J L. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 8.Compere S J, Palmiter R D. Cell. 1981;25:233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 9.Poirier L A. Drug Metab Rev. 1994;26:185–199. doi: 10.3109/03602539409029790. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y-I, Giuliano A, Hatch K D, Schneider A, Nour M A, Dallal G E, Selhub J, Mason J B. Cancer. 1994;74:893–899. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 11.Rossman T G, Stone D, Molina M, Troll W. Environ Mutagen. 1980;2:371–379. doi: 10.1002/em.2860020307. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson-Kram D, Montalbano D. Environ Mutagen. 1985;7:787–804. doi: 10.1002/em.2860070515. [DOI] [PubMed] [Google Scholar]
- 13.Gebhart E, Rossman T G. In: Metals and Their Compounds in the Environment. Merian E, editor. New York: VCH; 1991. pp. 617–641. [Google Scholar]
- 14.Lee T C, Oshimura M, Barrett J C. Carcinogenesis. 1985;6:1421–1426. doi: 10.1093/carcin/6.10.1421. [DOI] [PubMed] [Google Scholar]
- 15.Idoine J B, Elliott J M, Wilson M J, Weisburger E K. In Vitro. 1976;12:541–553. doi: 10.1007/BF02797437. [DOI] [PubMed] [Google Scholar]
- 16.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiraishi N, Hochadel J F, Coogan T P, Koropatnick J, Waalkes M P. Toxicol Appl Pharmacol. 1995;130:229–236. doi: 10.1006/taap.1995.1028. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J. Nucleic Acids Res. 1994;22:1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waalkes M P, Goering P L. Chem Res Toxicol. 1990;3:281–288. doi: 10.1021/tx00016a001. [DOI] [PubMed] [Google Scholar]
- 20.Eaton D L, Toal B F. Toxicol Appl Pharmacol. 1982;66:134–142. doi: 10.1016/0041-008x(82)90068-0. [DOI] [PubMed] [Google Scholar]
- 21.Adams R L P, Ranaldi A, Seivwright C. J Biochem Biophys Methods. 1991;22:19–22. doi: 10.1016/0165-022x(91)90077-a. [DOI] [PubMed] [Google Scholar]
- 22.Wagner J, Claverie N, Danzin C. Anal Biochem. 1984;140:108–116. doi: 10.1016/0003-2697(84)90140-4. [DOI] [PubMed] [Google Scholar]
- 23.Waalkes M P, Miller M S, Wilson M J, Bare R M, McDowell A E. Chem Biol Interact. 1988;66:189–204. doi: 10.1016/0009-2797(88)90071-3. [DOI] [PubMed] [Google Scholar]
- 24.Belinsky S A, Nikula K J, Baylin S B, Issa J-P J. Proc Natl Acad Sci USA. 1996;93:4045–4050. doi: 10.1073/pnas.93.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Counts J D, Goodman J I. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 26.Vahter M. Environ Res. 1981;25:286–293. doi: 10.1016/0013-9351(81)90030-x. [DOI] [PubMed] [Google Scholar]
- 27.Styblo M, Delnomdedieu M, Thomas D J. In: Toxicology of Metals: Biochemical Aspects. Goyer R A, Cherian M G, editors. Berlin: Springer; 1995. pp. 407–433. [Google Scholar]
- 28.Lengauer C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razin A, Cedar H. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holliday R. Biol Rev Cambridge Philos Soc. 1991;65:431–471. doi: 10.1111/j.1469-185x.1990.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 31.Holliday R. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 32.Ray J S, Harbison M L, McClain R M, Goodman J I. Mol Carcinog. 1994;9:155–166. doi: 10.1002/mc.2940090307. [DOI] [PubMed] [Google Scholar]
- 33.Loennechen T, Nilsen I W, Moens U, Andersen A, Aarbakke J. Biochem Pharmacol. 1992;44:1283–1289. doi: 10.1016/0006-2952(92)90527-p. [DOI] [PubMed] [Google Scholar]
- 34.Simile M M, Pascale R, De Miglio M R, Nufis A, Daino L, Seddaiu M A, Gaspa L, Feo F. Cancer Lett. 1994;79:9–16. doi: 10.1016/0304-3835(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 35.Gubits R M. Oncogene. 1988;3:163–168. [PubMed] [Google Scholar]
- 36.Wang, Z., Hou, G. & Rossman, T. B. (1994) Environ. Health Perspect. 102, Suppl. 3, 97–100. [DOI] [PMC free article] [PubMed]
- 37.Burden R H. Biochem J. 1986;240:313–324. doi: 10.1042/bj2400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin K V, Tanaka S, Darlington G, Pastan I, Gottesman M M. J Biol Chem. 1990;265:221–226. [PubMed] [Google Scholar]
- 39.Lee T C, Tanaka N, Lamb W P, Gilmer T M, Barrett J C. Science. 1988;241:79–81. doi: 10.1126/science.3388020. [DOI] [PubMed] [Google Scholar]
- 40.Schröter C, Parzefall W, Schröter H, Schulte-Hermann R. Cancer Res. 1987;47:80–88. [PubMed] [Google Scholar]
- 41.Wolff G C, Roberts D W, Morissey D W, Greenman D L, Allen R R, Campell W L, Bergman H, Nesnow S, Frith C H. Carcinogenesis. 1987;8:1889–1897. doi: 10.1093/carcin/8.12.1889. [DOI] [PubMed] [Google Scholar]
- 42.Levine A, Cantoni G L, Razin A. Proc Natl Acad Sci USA. 1991;88:6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salehi-Ashitiani K, Widrow R J, Markert C L, Goldberg E. Proc Natl Acad Sci USA. 1993;90:8886–8890. doi: 10.1073/pnas.90.19.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waalkes M P, Diwan B A, Weghorst C M, Ward J M, Rice J M, Cherian M G, Goyer R A. J Pharmacol Exp Ther. 1993;266:1656–1663. [PubMed] [Google Scholar]
- 45.Waalkes M P, Diwan B A, Rehm S, Ward J M, Moussa M, Cherian M G, Goyer R A. J Pharmacol Exp Ther. 1996;277:1026–1033. [PubMed] [Google Scholar]
- 46.Reynolds S H, Stowers S J, Maronpot R R, Anderson M W, Aaronson S A. Proc Natl Acad Sci USA. 1986;83:33–37. doi: 10.1073/pnas.83.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Counts J L, Goodman J I. Mol Carcinog. 1994;11:185–188. doi: 10.1002/mc.2940110402. [DOI] [PubMed] [Google Scholar]
- 48.Aposhian H V, Aposhian M M. J Am Coll Toxicol. 1989;8:1297–1305. [Google Scholar]
- 49.Crooks P A, Tribe M J, Pinney R J. J Pharm Pharmacol. 1984;36:85–89. doi: 10.1111/j.2042-7158.1984.tb02999.x. [DOI] [PubMed] [Google Scholar]
- 50.Bechthold A, Floss H G. Eur J Biochem. 1994;224:431–437. doi: 10.1111/j.1432-1033.1994.00431.x. [DOI] [PubMed] [Google Scholar]
- 51.Henning S M, McKee R W, Swendseid M E. J Nutr. 1989;119:1478–1482. doi: 10.1093/jn/119.10.1478. [DOI] [PubMed] [Google Scholar]