Abstract
In Escherichia coli K-12, 11 fla genes and a hag gene are located between his and uvrC, making two clusters at map positions 42.5 and 43.0 min. Nondefective transducing lambda phages for these genes were isolated. Low-frequency-transducing donors were constructed starting from lysogens of lambda cI857 in which the prophage is integrated at a secondary attachment site at 44 min on the E. coli map. Two strategies were used to delete the region between the prophage and the fla genes. Deletion mutants of the supD locus between fla and the prophage were isolated by selecting for loss of Su1+, an allele of supD. A strain with a deletion starting within the prophage and ending at a position close to the fla genes was isolated from heat-resistant derivatives of the lysogen. A lysogen of lambda b2 was then constructed in which the prophage had integrated at the site of the defective prophage by means of recombination with residual lambda deoxyribonucleic acid. From low-frequency-transducing lysate of the donor strains thus constructed, either directly or in combination with a procedure that extends the loci transduced, various lambda pfla's were isolated. lambda pflaL1 carries all nine fla genes at 43 min, and lambda pflaH14 carries hag and two fla genes at 42.5 min.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- APPLEYARD R. K., MCGREGOR J. F., BAIRD K. M. Mutation to extended host range and the occurrence of phenotypic mixing in the temperate coliphage lambda. Virology. 1956 Aug;2(4):565–574. doi: 10.1016/0042-6822(56)90012-5. [DOI] [PubMed] [Google Scholar]
- Adler J. Chemotaxis in bacteria. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- Andoh T., Ozeki H. Suppressor gene Su3+ of E. coli, a structural gene for tyrosine TRNA. Proc Natl Acad Sci U S A. 1968 Mar;59(3):792–799. doi: 10.1073/pnas.59.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Purification of intact flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):376–383. doi: 10.1128/jb.105.1.376-383.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmitt K., Simon M. Purification and thermal stability of intact Bacillus subtilis flagella. J Bacteriol. 1971 Jan;105(1):369–375. doi: 10.1128/jb.105.1.369-375.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A., Celis J. E. Mischarging single and double mutants of Escherichia coli sup3 tyrosine transfer RNA. J Mol Biol. 1974 Mar;83(3):333–351. doi: 10.1016/0022-2836(74)90283-6. [DOI] [PubMed] [Google Scholar]
- Iino T. Genetics and chemistry of bacterial flagella. Bacteriol Rev. 1969 Dec;33(4):454–475. doi: 10.1128/br.33.4.454-475.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. I. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965 Nov;14(1):85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- Komeda Y., Suzuki H., Ishidsu J. I., Iino T. The role of cAMP in flagellation of Salmonella typhimurium. Mol Gen Genet. 1976 Dec 31;142(4):289–298. doi: 10.1007/BF00271253. [DOI] [PubMed] [Google Scholar]
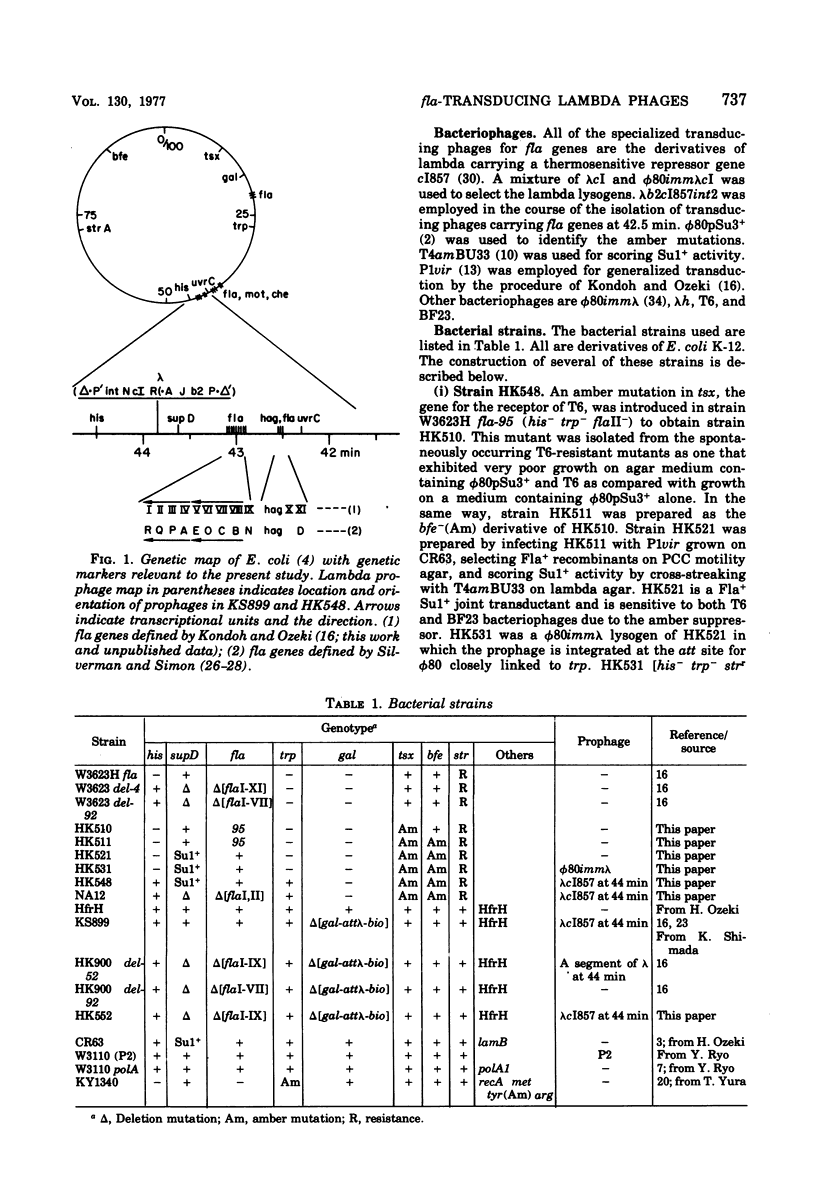
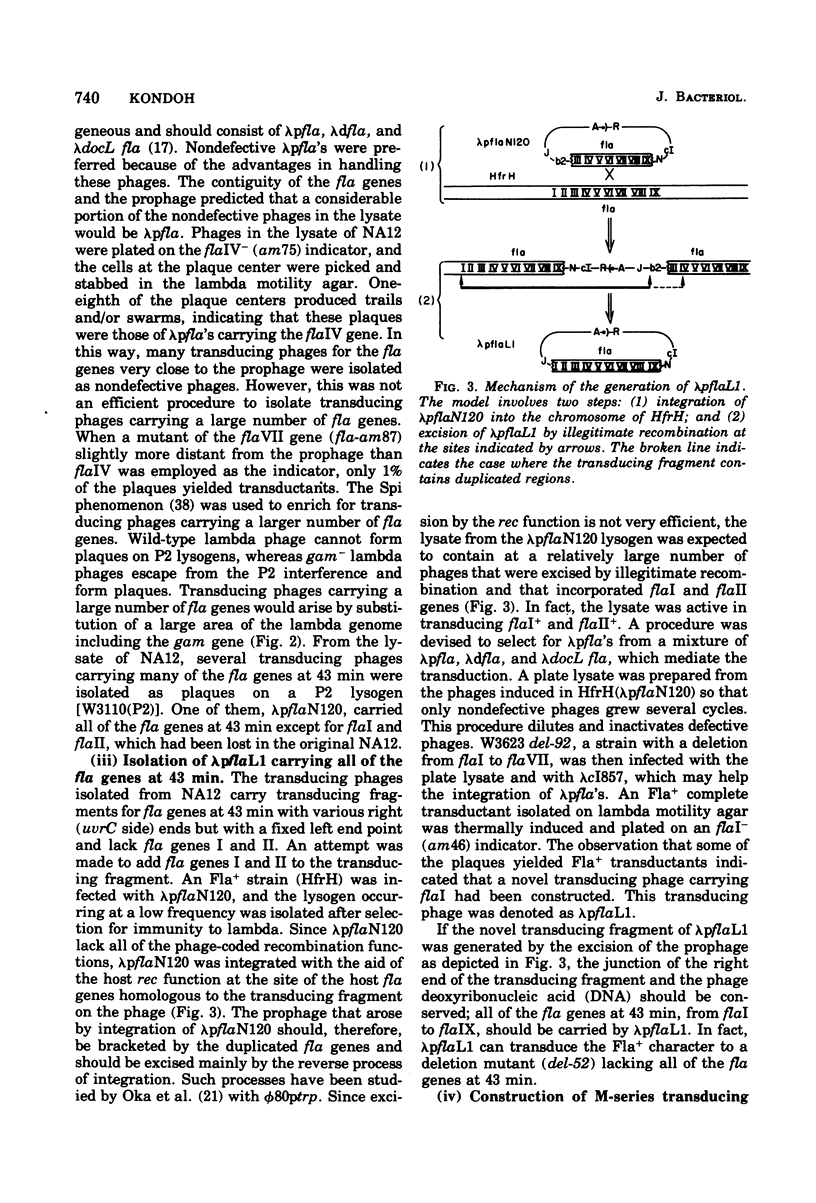
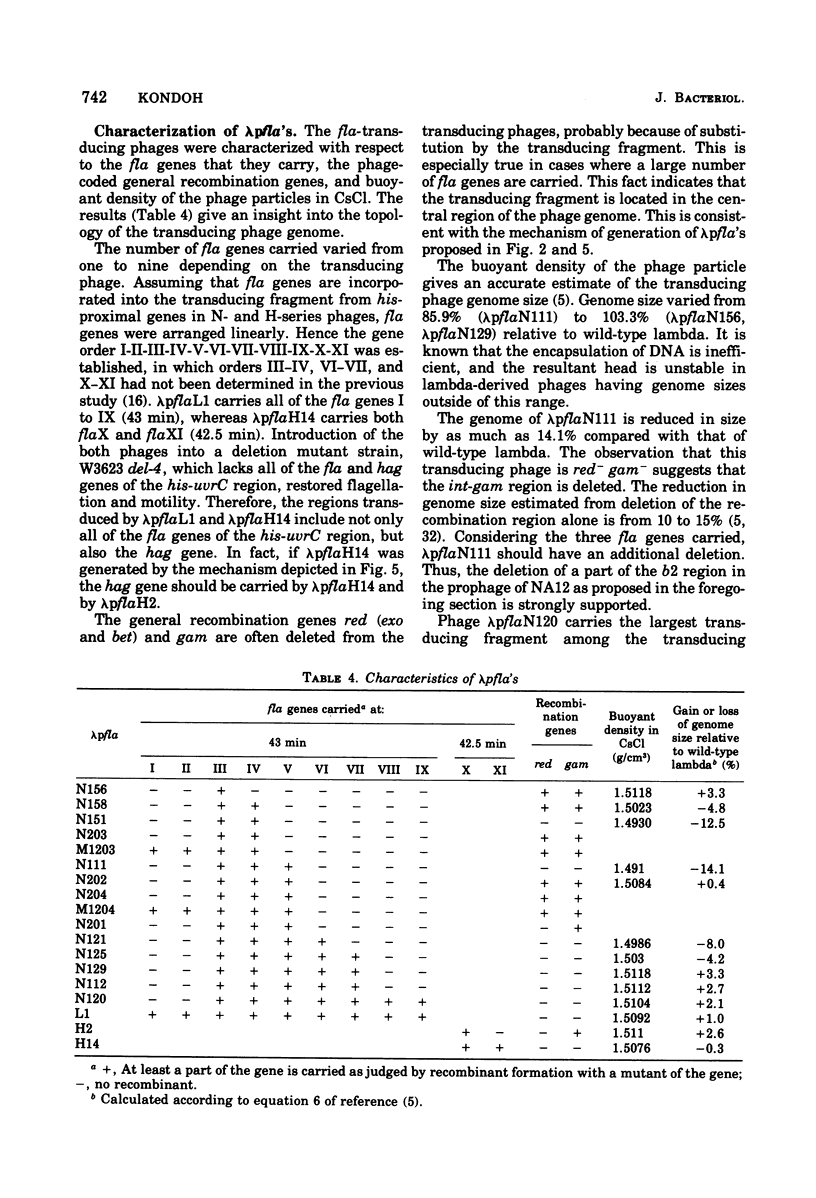
- Kondoh H., Ozeki H. Deletion and amber mutants of fla loci in Escherichia coli K-12. Genetics. 1976 Nov;84(3):403–421. doi: 10.1093/genetics/84.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Horiuchi T. Isolation and characterization of a temperature-sensitive amber suppressor mutant of Escherichia coli K12. Mol Gen Genet. 1973;123(1):77–88. doi: 10.1007/BF00282991. [DOI] [PubMed] [Google Scholar]
- Oka A., Ozeki H., Inselburg J. Integration and excision of phi-80pt prophage in Escherichia coli. I. Replacement of tryptophan genes of phi-80pt with the host alleles through the lysogenic process. Virology. 1971 Dec;46(3):556–566. doi: 10.1016/0042-6822(71)90059-6. [DOI] [PubMed] [Google Scholar]
- SUSSMAN R., JACOB F. [On a thermosensitive repression system in the Escherichia coli lambda bacteriophage]. C R Hebd Seances Acad Sci. 1962 Feb 19;254:1517–1519. [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Silerman M., Matsumura P., Draper R., Edwards S., Simon M. I. Expression of flagellar genes carried by bacteriophage lambda. Nature. 1976 May 20;261(5557):248–250. doi: 10.1038/261248a0. [DOI] [PubMed] [Google Scholar]
- Silverman M., Matsumura P., Simon M. The identification of the mot gene product with Escherichia coli-lambda hybrids. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3126–3130. doi: 10.1073/pnas.73.9.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J Bacteriol. 1974 Dec;120(3):1196–1203. doi: 10.1128/jb.120.3.1196-1203.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Genetic analysis of bacteriophage Mu-induced flagellar mutants in Escherichia coli. J Bacteriol. 1973 Oct;116(1):114–122. doi: 10.1128/jb.116.1.114-122.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Genetic analysis of flagellar mutants in Escherichia coli. J Bacteriol. 1973 Jan;113(1):105–113. doi: 10.1128/jb.113.1.105-113.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Positioning flagellar genes in Escherichia coli by deletion analysis. J Bacteriol. 1974 Jan;117(1):73–79. doi: 10.1128/jb.117.1.73-79.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Iino T. Absence of messenger ribonucleic acid specific for flagellin in non-flagellate mutants of Salmonella. J Mol Biol. 1975 Jul 15;95(4):549–556. doi: 10.1016/0022-2836(75)90316-2. [DOI] [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. Physical mapping of the att-N region of coliphage lambda: apparent oversaturation of coding capacity in the gam-ral segment. Biochimie. 1974;56(11-12):1497–1503. doi: 10.1016/s0300-9084(75)80272-0. [DOI] [PubMed] [Google Scholar]
- Vary P. S., Stocker B. A. Nonsense motility mutants in Salmonella typhimurium. Genetics. 1973 Feb;73(2):229–245. doi: 10.1093/genetics/73.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H., Inokuchi H., Ozeki H. Excision and duplication of su3+-transducing fragments carried by bacteriophage phi 80. I. Novel structure of phi 80sus2psu3+ DNA molecule. J Virol. 1976 Jun;18(3):1016–1023. doi: 10.1128/jvi.18.3.1016-1023.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H., Inokuchi H., Ozeki H. Excision and duplication of su3+-transducing fragments carried by bacteriophage theta80. II. Red- or Rec-dependent excision and duplication. J Mol Biol. 1976 Sep 5;106(1):133–150. doi: 10.1016/0022-2836(76)90304-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Iino T., Horiguchi T., Ota K. Genetic analysis of fla and mot cistrons closely linked to H1 in Salmonella abortusequi and its derivatives. J Gen Microbiol. 1972 Apr;70(1):59–75. doi: 10.1099/00221287-70-1-59. [DOI] [PubMed] [Google Scholar]



