Abstract
The small subunit of calpain, a calcium-dependent cysteine protease, was found to interact with the cytoplasmic domain of the common cytokine receptor γ chain (γc) in a yeast two-hybrid interaction trap assay. This interaction was functional as demonstrated by the ability of calpain to cleave in vitro-translated wild-type γc, but not γc containing a mutation in the PEST (proline, glutamate, serine, and threonine) sequence in its cytoplasmic domain, as well as by the ability of endogenous calpain to mediate cleavage of γc in a calcium-dependent fashion. In T cell receptor-stimulated murine thymocytes, calpain inhibitors decreased cleavage of γc. Moreover, in single positive CD4+ thymocytes, not only did a calpain inhibitor augment CD3-induced proliferation, but antibodies to γc blocked this effect. Finally, treatment of cells with ionomycin could inhibit interleukin 2-induced STAT protein activation, but this inhibition could be reversed by calpain inhibitors. Together, these data suggest that calpain-mediated cleavage of γc represents a mechanism by which γc-dependent signaling can be controlled.
Mutation of the common cytokine receptor γ chain, γc, results in X-linked severe combined immunodeficiency in humans (1, 2). The severity of this disease is explained by the participation of γc as an essential component of the receptors for interleukin (IL) 2, IL-4, IL-7, IL-9, and IL-15 (3–9). The phenotype in humans with X-linked severe combined immunodeficiency (2) and in γc-deficient mice (10–12) indicates the vital role played by γc in intrathymic T cell development. In normal mice, γc mRNA can be detected as early as day 13.5 in fetal thymus (C. Sommers and P. E. Love, personal communication) and in all subpopulations of mature thymocytes (13), consistent with vital role(s) for γc throughout thymic development. In addition to its roles in cytokine signaling and thymic development, signaling through γc has been proposed to prevent T cell anergy, based on observations that after T cell receptor (TCR)-mediated signaling, IL-2, IL-4, and IL-7 each can prevent the induction of anergy (14).
For the IL-2 receptor, heterodimerization of the cytoplasmic regions of γc and IL-2Rβ is necessary and sufficient for signaling (15, 16). IL-2Rβ has been reported to interact with multiple signaling molecules, including Lck (17), Shc (18, 19), Syk (20), PI 3-kinase (21), Jak1 (8, 14, 22), and STAT proteins (19, 23). In contrast, the only signaling molecule known to interact with γc is the Janus family tyrosine kinase, Jak3 (8, 14, 22). We now report that the small subunit of the calcium-activated neutral protease, calpain, an enzyme implicated in T cell apoptosis (24–26), can associate with γc. We show that calpain can cleave γc, that anti-CD3 treatment of freshly isolated thymocytes resulted in proteolysis of γc, and that calpain-specific inhibitors protected against this proteolysis and could augment anti-CD3-induced proliferation. Moreover, ionomycin could inhibit IL-2-induced STAT protein activation, and calpain inhibitors reversed this effect. Proteolysis of γc by calpain therefore may represent a mechanism of controlling levels of γc, thereby modulating cytokine-dependent signaling responses.
MATERIALS AND METHODS
Yeast Two-Hybrid System.
To generate the “bait plasmid” (pAS-CYH2-γc), the human γc cytoplasmic domain was amplified by PCR using sense (5′-ATCATCATCCATGGTGTATTTCTGGCTGGAACG-3′; +835 to +854) and antisense (5′-ATCATCATGGATCCATCGGTTCAGGAACAATCGG-3′; +1,409 to +1,389) primers containing NcoI and BamHI sites, respectively (underlined), and subcloned between the NcoI and BamHI sites of pAS1-CYH2, allowing production in yeast of a Gal4 binding domain/γc cytoplasmic domain fusion protein. Y190 cells (MATa gal4 gal80 his3 trp1–901 ade2–101 ura3–52 leu2–3,-112 + URA3::GAL–>lacZ, LYS2::GAL(UAS)–>HIS3 cyhr) were transformed to Trp prototrophy with pAS1-CYH2-γc. Y190 cells contain two reporter genes, lacZ and HIS3, under Gal4 control, allowing the identification of cDNA clones encoding interacting proteins by β-galactosidase (β-gal) production and by the growth of transformed cells on His− medium in the presence of 3-amino-1,2,4-triazole (27). After confirming that pAS1-CYH2-γc by itself did not direct transcription of β-gal, Y190 (pAS1-CYH2-γc) cells were transformed by the lithium acetate method using denatured sheared herring testes DNA as a carrier and approximately 107 clones from a cDNA library in which human Epstein–Barr virus-transformed B cell cDNAs were fused to the Gal4 activation domain in pACT (27). Cells were plated on Leu−, Trp−, and His− plates with 25 mM 3-amino-1,2,4-triazole (Sigma) and incubated for up to 8 days at 30°C. His+ colonies were evaluated for β-gal activity using a filter-lift assay. After the bait construct was eliminated by plating on Leu− plates containing cycloheximide (27), 347 β-gal+ clones were mated with Y187 yeast (MATα gal4 gal80 his3 trp1–901 ade2–101 ura3–52 leu2–3,-112 met− URA3::GAL–>lacZ) carrying pAS1-CYH2 without an insert or with SNF-1 or p53 as fusion proteins with the Gal4 activation domain (negative control) or with the bait construct (positive control). The β-gal assay was used to eliminate nonspecific interactions, and 21 cDNA plasmids were rescued from yeast and transformed into bacteria. Plasmid DNAs were isolated and sequenced using either the prism ready reaction dyedeoxy terminator cycle sequencing kit (Applied Biosystems) or Sequenase.
In Vitro Translation of γc and Cleavage with Calpain.
In vitro translation reactions were performed using the TNT coupled wheat germ extract system (Promega), [35S]methionine (Amersham), T7 polymerase, and γc constructs cloned in pAlter (Promega). Each reaction (50 μl) was passed through a Sephadex G-50 column (5 Prime → 3 Prime), and 10 μl of in vitro-translated protein product was mixed with 5 μl of m-calpain (Sigma) solution (final concentrations 0–25 μg/ml) on ice. Calpain was activated by adding 5 μl of 400 mM Tris⋅HCl, pH 7.5/24 mM CaCl2/2 mM DTT at 30°C. After 30 min, reactions were stopped with 20 μl of 2× SDS sample buffer, run on SDS gels (NOVEX, San Diego), and analyzed with a PhosphorImager (Molecular Dynamics).
Mutation of the PEST (Proline, Glutamate, Serine, and Threonine) Sequence in γc.
The PEST sequence in γc (KGLAESLQPDYSE) was mutated to KGLAVGLQLDYSE using the Altered Sites II in vitro mutagenesis system (Promega) and an oligonucleotide (5′-GGTGTGTCTAAGGGACTGGCTGTGGGTCTGCAGCTAGACTACAGTGAACGACTCTGC-3′).
Cleavage of γc in YT Cell Lysates and Intact Cells.
YT cells were washed with calcium-free PBS, pH 7.4 and lysed using Brij 96 lysis buffer (10 mM Tris, pH 7.5 containing 150 mM NaCl, 0.875% Brij96, and 0.125% Nonidet-P 40) without protease inhibitors, EDTA, or EGTA, for 20 min on ice. Lysates were centrifuged at 15,000 rpm for 20 min at 4°C and incubated with CaCl2 (final concentration 5 mM) or EGTA (final concentration 10 mM) at 37°C for 0 to 60 min. To determine the specificity of proteolysis by calpain, calpastatin (20 μM, Calbiochem), an endogenous protease inhibitor that acts specifically on calpain, or antipain [S-(1 carboxy-2-phenylethyl)-l-carbamyl=l-arginyl-l-valylargininal] (Calbiochem), another potent inhibitor of calpain, were incubated in the presence of 5 mM CaCl2. The reaction was terminated with EGTA (10 mM final concentration) and the protease inhibitor mix. Immunoprecipitations were performed with immobilized anti-chicken IgY (Promega) and either chicken anti-human γc antibody (Promega) or chicken IgY (H+Y) as a control antibody (Promega). After 3 hr at 4°C, immunoprecipitates were extensively washed in the same lysis buffer and analyzed on 10–20% Tricine SDS gels (NOVEX). Immunoblotting was carried out with R878 antiserum to the cytoplasmic tail of γc (5, 6) and developed using ECL (Amersham).
For experiment in intact YT cells, cells were washed twice with calcium-free PBS and incubated for 30, 60, or 90 min with 5 μM ionomycin (Sigma) and either 2.5 mM CaCl2 or 0.5 mM EGTA in calcium-free PBS. Cells were lysed with Brij96 lysis buffer containing 5 mM EGTA and 2.5 mM EDTA and a protease inhibitor mix containing 10 μg/ml leupeptin (Boehringer Mannheim), 240 μg/ml 4-[2-aminoethyl]-benzenesulfonyl fluoride hydrochloride (ICN), and 10 μg/ml aprotinin (ICN). Lysates were immunoprecipitated with anti-γc antibody (Promega) and run on SDS gels (NOVEX). Gels were immunoblotted with R878 antiserum to γc and developed using ECL. Cell lysates also were analyzed on 7.5% tris-glycine SDS gels and stained with Coomassie blue to evaluate nonspecific degradation during the incubation. To evaluate the dose-responsiveness of ionomycin on γc cleavage, YT cells were treated with 0, 2, 20, or 40 μM ionomycin, and γc cleavage was evaluated as above. To confirm that IL-2 does not induce cleavage of γc, YT cells were treated with 2 nM IL-2 in PBS containing 2.5 mM CaCl2 for 60 min at 37°C, and γc cleavage was evaluated by Western blotting with R878 antiserum.
TCR-Mediated γc Proteolysis in Thymocytes.
Murine thymocytes from 4-week-old C57BL/6 mice were washed twice with calcium-free PBS and resuspended at 107 cells/ml in RPMI medium 1640 containing 10% fetal bovine serum (FBS). Cells were preincubated for 15 min with 0 or 25 μM AllnM (calpain inhibitor II; Calbiochem), a potent inhibitor of calpain (25). Cells then were incubated with 20 μg/ml of purified hamster anti-mouse CD3ɛ mAb (145–2C11, PharMingen) in RPMI medium 1640/10% FBS for 15 min, and then crosslinked using 60 μg/ml of affinity-purified, unconjugated goat anti-hamster IgG (GAHIgG, Jackson Immunoresearch) (29) for the indicated periods of time. Cells were lysed at 5 × 107 cells/ml in EB-1 lysis buffer (0.5% Nonidet-P 40/140 mM NaCl/50 mM Tris, pH 7.5) containing 5 mM EGTA, 5 mM EDTA, and the protease inhibitor mix. Lysates were run on 4–20% or 4–12% Tris SDS/polyacrylamide gels (NOVEX) and immunoblotted with R878 antiserum.
Cleavage of Flag-Tagged γc by Thymocyte Lysates.
γc was cloned into the pFLAG-CMV-1 vector (Kodak) and transfected into 293 T cells by calcium phosphate (5 Prime → 3 Prime). The resulting FLAG-γc protein was purified by anti-FLAG antibody (M2)-conjugated affinity beads (Kodak). Total thymocytes from 4-week-old C57BL/6 mice were treated with goat-anti-hamster IgG (GAHIgG, PharMingen), anti-CD3ɛ + GAHIgG. Cells were lysed with EB-1 lysis buffer lacking protease inhibitors and calcium chelators or containing calpastatin. The purified FLAG-tagged γc was incubated with the indicated cell lysates or with 0.1 μM purified m-calpain (Calbiochem 208715) for 30 min at 37°C. Lysates then were run on SDS gels and analyzed by Western blotting with anti-FLAG M2 antibody (Kodak) using ECL.
Isolation of CD4+ Thymocytes.
Thymocytes (400 × 106) from C57BL/6 mice (The Jackson Laboratory) in 1 ml of Hanks’ balanced salt solution (HBSS) were incubated with an equal volume of peanut agglutinin (0.5 mg/ml, Sigma) for 10 min at room temperature (25). Cells then were loaded onto 10 ml of 2% BSA in HBSS and allowed to stand for 30 min at room temperature. CD4+ cells were purified from the cells at the interface by complement-mediated depletion of Ia+ and CD8+ cells using anti-Ia mAb M5114 and anti-CD8 mAb 3.155.
IL-7-Dependent Protection Against Apoptosis in Thymocytes.
Total thymocytes from 4-week-old C57BL/6 mice, of which 91% were CD4+CD8+ double positive cells, were resuspended at 2 × 106/ml in RPMI medium 1640 containing 10% charcoal-treated FBS (Cocalico Biologicals, Reamstown, PA), and incubated for 30 min at 37°C with 400 nM calpastatin. Cells were transferred at 2× 105 cells/well in 100 μl to 96-well plates (Costar) that were precoated overnight at 4°C with 30 μl of 50 μg/ml of both anti-CD3 and anti-CD28 (375.1 mAb) (30) or with purified polyclonal hamster IgG (PharMingen) in 0.1 M Tris, pH 9.5. After 1 hr (37°C, 5% CO2), 20 ng/ml recombinant murine IL-7 (R & D Systems) was added. Cells were harvested 18 hr later and stained with ethidium bromide, and 104 cells/condition were analyzed by FACSort, as described (31). The % viable (cells not staining) and nonviable cells then was determined.
Electrophoretic Mobility Shift Assays.
Human peripheral blood lymphocytes were isolated from normal donors, stimulated with 1 μg/ml of phytohemagglutinin-L (Boehringer Mannheim) for 48 hr, washed, and rested overnight in medium lacking phytohemagglutinin. Cells were incubated for 30 min in RPMI medium 1640 (without calcium nitrate, GIBCO/BRL) containing 10% FBS and 2.5 mM CaCl2 with or without 20 μM purified human calpastatin domain I peptide (amino acids 184–217, synthesized by M. Berne, Tufts University) (32) or EGTA. Where indicated, 4 μM ionomycin was added for 30 min at 37°C with gentle agitation. Cells then were stimulated with 1.3 nM human IL-2 for 15 min at 37°C, lysed in 20 mM Hepes, pH 7.8/450 mM NaCl/0.4 mM EDTA/2.5 mM EGTA/0.5 mM DTT/25% glycerol/0.5 mM 4-[2-aminoethyl]-benzenesulfonyl fluoride hydrochloride/10 μg/ml leupeptin/10 μg/ml aprotinin by the freeze-thaw method, and electrophoretic mobility shift assays were performed (23) using an end-labeled double-stranded β-casein probe (5′-AGCTTAGATTTCTAGGAATTCG-3′) and 15 μg of protein (determined using the Bio-Rad Protein Assay kit) in each lane.
RESULTS
Association of γc and Calpain in the Yeast Two-Hybrid System.
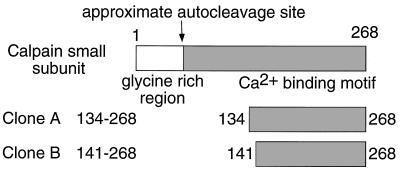
To identify additional proteins capable of associating with the cytoplasmic domain of γc, we used a yeast two-hybrid method (27, 33) in which the entire γc cytoplasmic domain was used as bait (see Material and Methods). Of 107 cDNA clones screened, 21 specifically interacting clones were identified, two of which encoded amino acids 134–268 (clone A) and amino acids 141–268 (clone B), respectively, of the small subunit of human calpain (Fig. 1).
Figure 1.
The small subunit of calpain associates with γc. Using the yeast two-hybrid system with the γc cytoplasmic domain as bait, two cDNAs were identified that corresponded to the C-terminal region of the small subunit of calpain distal to the autoproteolytic cleavage site (35).
Calpains are calcium-activated neutral proteases that are expressed in many mammalian tissues (34–36), including lymphocytes (37), and can catalyze the cleavage of a wide range of proteins, including enzymes, transcription factors, cytoskeletal proteins, and members of receptor signaling pathways (34–36). Each calpain has a unique 80-kDa large subunit but shares a common 30-kDa small subunit (34–36). Calpain is converted from an inactive proenzyme to its active form by autoproteolytic cleavage of both subunits, with cleavage of the small subunit to a 17-kDa C-terminal fragment that remains associated with the large subunit (35). As the clones we identified in the two-hybrid analysis were C terminally biased, our data suggest that autoproteolytically cleaved active calpain can interact with γc.
Cleavage of γc by Calpain.
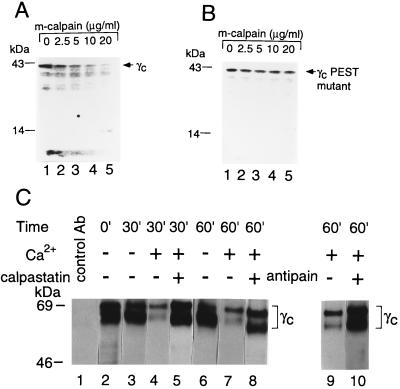
Efforts to coprecipitate calpain and γc yielded at best low levels of coprecipitation (data not shown). However, physiologically relevant interactions can potentially either be too transient or of insufficient affinity to allow efficient coprecipitation. Furthermore, it was conceivable that efficient interaction might occur only after calpain activation and that such activation might cleave γc, decreasing our ability to observe the interaction by coprecipitation. Therefore, we investigated the ability of calpain to cleave γc. Proteins that are targets for calpain typically have a region rich in proline (P), glutamic acid (E), serine (S), and threonine (T) flanked by clusters of positively charged amino acids (28, 38, 39). Using the pest-find computer program (28), we found such a region beginning in the SH2-subdomain homology region (3) of the human γc cytoplasmic domain. This sequence (GLAESLQPDYSE; pest score = −3.95, scores > −5 are significant), which is flanked by positively charged amino acids, was conserved in canine γc, and an even higher pest score was found in murine γc (GLTESLQPDYSE; pest score = +0.59). Although PEST sequences are not necessarily required for calpain-mediated cleavage of substrates (40), it was striking that calpain could cleave in vitro-translated wild-type γc (Fig. 2A), but not γc in which the PEST region in human γc was mutated from GLAESLQPDYSE to GLAVGLQLDYSE (which does not score as a valid PEST sequence) (Fig. 2B).
Figure 2.
Cleavage of γc by calpain. In vitro-translated wild-type human γc (A) and γc in which the PEST sequence was mutated (B) were treated with m-calpain and run on 4–20% SDS gels. Although mature γc is approximately 64 kDa, in vitro-translated γc migrates at approximately 42 kDa, at least in part due to the lack of glycosylation. We confirmed a report that in vitro-translated c-Fos is sensitive to cleavage by calpain, whereas chloramphenicol acetyltransferase is not (65). Murine wild-type and PEST-mutated γc yielded similar results to those shown for human γc (data not shown). (C) Proteolysis of γc by calpain in YT cells. YT cell lysates were incubated with 5 mM CaCl2 (lanes 4, 5, and 7–10) or 10 mM EGTA + 2.5 mM EDTA (lanes 3 and 6) at 37°C for 0–60 min, and reactions were stopped by the addition of 10 mM EGTA/10 μg/ml leupeptin (Boehringer Mannheim)/240 μg/ml 4-[2-aminoethyl]-benzenesulfonyl fluoride hydrochloride/10 μg/ml aprotinin (ICN). In lanes 4, 7, and 10, 20 μM calpastatin or 50 μg/ml antipain also were added. Reactions were stopped with 10 mM EGTA and the protease inhibitor mix. Lysates were immunoprecipitated with chicken anti-human γc (lanes 2–10) or control chicken IgY (lane 1), and immunoblotted using R878 antiserum to γc. As R878 antiserum recognizes the C-terminal end of γc, cleavage in the cytoplasmic domain would result in immunoreactive fragments too small to be retained on these gels. The cleavage was specific, as shown by the lack of general degradation of cellular proteins as detected by Coomassie staining (data not shown).
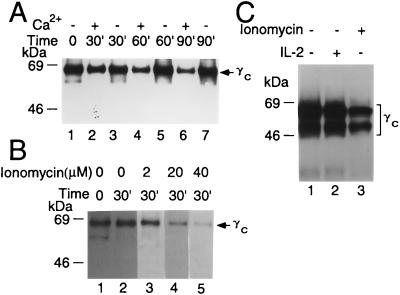
We next evaluated calcium-dependent cleavage of γc in YT cell lysates. YT cells were washed with calcium-free PBS and lysed with Brij 96 lysis buffer. Lysates were incubated with 5 mM CaCl2 or 10 mM EGTA + 2.5 mM EDTA for 0 to 60 min. Reactions were stopped by the addition of EGTA (final concentration 10 mM) and protease inhibitors, and lysates were immunoprecipitated with chicken anti-human γc antibody (Fig. 2C, lanes 2–10) or control IgY (Fig. 2C, lane 1), run on SDS gels, and immunoblotted with R878 antiserum to γc. The γc bands were diminished in intensity only in the presence of 5 mM CaCl2 (Fig. 2C, lanes 4 and 7 vs. lanes 3 and 6). Cleavage was inhibited by calpastatin (Fig. 2C, lanes 5 and 8), a specific calpain inhibitor (32, 34), and by antipain (Fig. 2C, lane 10 vs. 9), another potent inhibitor of calpain (41). These data indicate that calpain was the enzyme-mediating cleavage of γc. In addition to this calcium-activated cleavage of γc in YT cell lysates, treatment of intact YT cells with ionomycin also induced γc cleavage in a time-dependent manner (Fig. 3A). Cleavage was detected only in the presence of calcium (Fig. 3A) and was increased at high concentrations of ionomycin (Fig. 3B). As expected, cleavage of γc was not seen in response to IL-2 (Fig. 3), a stimulus that does not increase intracellular calcium concentrations (42).
Figure 3.
Cleavage of γc in YT cells in a time- and dose-dependent fashion. (A) Treatment with ionomycin induces cleavage of γc in a dose time-dependent manner in the presence (lanes 2, 4, and 6) but not the absence of calcium (lanes 3, 5, and 7). (B) Cleavage of γc increased with increasing levels of ionomycin. (C) Cleavage of γc by ionomycin but not by IL-2.
TCR Activation Mediated Proteolysis of γc in Thymocytes.
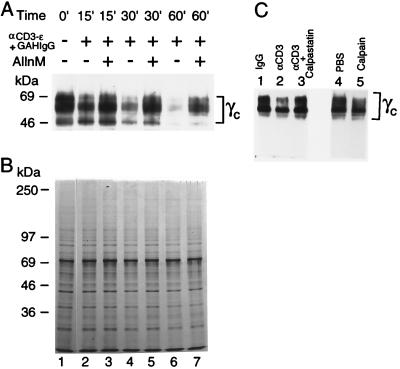
As noted above, IL-2 does not induce an increase in intracellular calcium and did not induce cleavage of γc. However, signaling through the TCR rapidly increases intracellular calcium (28, 43–46), leading us to hypothesize that TCR-mediated calcium flux might be sufficient to activate calpain and induce cleavage of γc. As X-linked severe combined immunodeficiency patients and γc-deficient mice exhibit greatly diminished thymic development, indicating the importance of γc in this process, we studied the possible role of γc cleavage in thymocytes. We first used anti-CD3 stimulation of murine thymocytes, a treatment known to induce apoptosis (47), as a way of increasing intracellular calcium concentrations and found that such treatment diminished the intensity of γc bands in a time-dependent manner (Fig. 4A, lanes 2, 4, and 6 vs. 1). Moreover, γc cleavage was inhibited when cells were preincubated with 25 μM AllnM (Fig. 4A, lanes 3, 5, and 7 vs. 2, 4, and 6), a potent inhibitor of m-calpain (34–36), indicating that γc was being cleaved by calpain. In contrast, anti-CD3 treatment did not induce a general degradation of cellular lysate proteins, as detected by Coomassie staining (Fig. 4B). To further confirm the activation of calpain in thymocytes, 293 T cells were transfected with a FLAG-tagged γc, and anti-FLAG immunoprecipitated protein was added to lysates from thymuses stimulated with control IgG or anti-CD3 in the presence vs. absence of calpastatin. Cleavage was seen with anti-CD3 and inhibited by calpastatin (Fig. 4C, lanes 2 and 3), again implicating calpain in the cleavage. As expected the FLAG-γc protein was cleaved by purified calpain, but not by PBS (Fig. 4C, lane 5 vs. 4).
Figure 4.
TCR-mediated proteolysis of γc in thymocytes. (A) Thymocytes from 4-week-old C57BL/6 mice were washed twice with calcium-free PBS and resuspended at 107 cells/ml in RPMI medium 1640 containing 10% FBS. Cells were preincubated for 15 min with 0 or 25 μM AllnM (lanes 3, 5, and 7), then incubated with 20 μg/ml of purified hamster anti-mouse CD3ɛ mAb (145–2C11, PharMingen) in RPMI medium 1640/10% FBS for 15 min, and crosslinked using 60 μg/ml of affinity-purified, unconjugated goat anti-hamster IgG (GAHIgG) for the indicated periods of time. Cell lysates were immunoblotted with R878 antiserum. The multiple forms of γc detected may include differentially glycosylated mature forms as well as precursor forms (66, 67). Because R878 recognizes only the C-terminal end of the cytoplasmic domain of γc, only fragments retaining the epitope are visualized. (B) Coomassie staining of the cellular lysates. (C) Cleavage of γc by calpain contained in thymic extracts. FLAG-γc protein was immunoprecipitated from transfected 293 T cells and then added to lysates from thymocytes treated with IgG (lane 1) or anti-CD3 (lanes 2 and 3). In lane 3, cells were lysed in the presence of calpastatin. As controls, γc-FLAG protein was incubated with either PBS (lane 4) or purified m-calpain (lane 5).
Calpain Inhibitors Minimally Augmented IL-7-Dependent Protection from Apoptosis Induced by TCR Stimulation in Thymocytes.
Cytokines that signal via γc, including IL-2, IL-4, and IL-7, can protect against apoptosis in lymphocytes by acting as survival factors (48–51). We therefore asked whether TCR-mediated cleavage of γc might influence cell viability after TCR stimulation. A calpastatin peptide alone did not significantly increase survival in thymocytes (consistent with previous observations; see ref. 25), but had a modest but reproducible positive effect on the ability of IL-7 to enhance cell survival (Table 1). However, the degree of protection did not achieve statistical significance and its physiological relevance remains unclear. The fact that IL-7 + calpastatin had such a modest effect may reflect incomplete inhibition of calpain by calpastatin or that calpain- and γc-independent mechanisms are primarily involved. Another inhibitor of calpain, AllnM, also exhibited similarly weak (and not statistically significant) effects on the ability of IL-7 to confer protective effects against anti-CD3-induced apoptosis (data not shown).
Calpain Inhibitors Increased Anti-CD3-Induced Proliferation and Reversed Ionomycin-Mediated Decreases in IL-2-Induced Stat5 Activation.
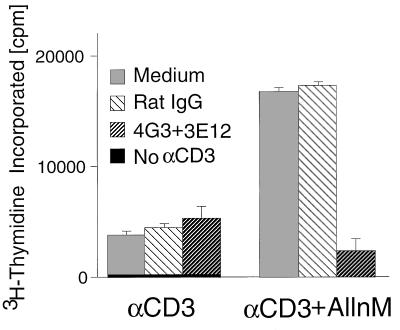
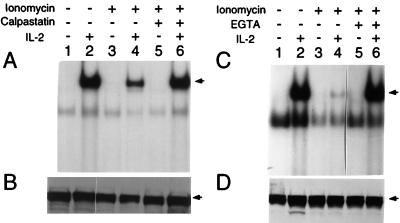
In contrast to the minor effect of calpastatin inhibitors on apoptosis, AllnM greatly increased anti-CD3-induced proliferation of CD4+ thymocytes (consistent with a previous report, ref. 25), but anti-γc antibodies that block cytokine binding (52) inhibited this effect (Fig. 5). These data indicate that activated calpain can inhibit the immunological response of cytokine(s) whose receptor(s) contain γc. Finally, we examined the effect of calpain inhibitors on STAT protein activation (Fig. 6). STAT protein DNA binding activity is induced in phytohemagglutinin-activated human peripheral blood lymphocytes stimulated with IL-2 (Fig. 6A, lane 2). This was diminished by the addition of ionomycin (to increase intracellular calcium) (Fig. 6A, lane 4), but the effect was reversed by calpastatin (Fig. 6A, lane 6) or EGTA (Fig. 6C, lane 6 vs. 4). Together, these data support the hypothesis that calpain can modulate γc-dependent signaling and suggest calpain may affect proliferation and STAT protein activation.
Figure 5.
Antibodies to γc block AllnM-mediated augmentation of anti-CD3-induced proliferation of CD4+ single positive thymocytes. CD4+ thymocytes were isolated from total thymocytes (see Materials and Methods) and cultured at 105 cells/well in a 96-well microtiter plate coated with anti-CD3 (1 μg/ml of 2C11 mAb) in the presence of 25 μM AllnM and 10 μg/ml of the indicated antibodies (no azide, low endotoxin, PharMingen). After 36 hr of culture, 0.5 μCi of [3H]thymidine was added to each well, and the wells were harvested 18 hr later. The data points are the mean ± SEM from three replicates.
Figure 6.
Inhibition of IL-2-induced STAT protein activation by ionomycin is reversed by EGTA or calpastatin. (A) Electrophoretic mobility shift assay using the β-casein probe and extracts from phytohemagglutinin blasts that were not stimulated or stimulated with IL-2 in the presence or absence of ionomycin and calpastatin, as indicated. (C) Electrophoretic mobility shift assay using the β-casein probe and extract from phytohemagglutinin blasts not stimulated or stimulated with IL-2 in the presence or bsence of ionomycin and EGTA, as indicated. (B and D) Western blotting of the lysates in A and C, respectively, was performed with anti-Stat5 (Transduction Laboratories, Lexington, KY).
DISCUSSION
We have demonstrated that the small subunit of calpain, a calcium-activated neutral protease, can physically associate with the γc cytoplasmic domain. This association was functionally important based on several lines of evidence. First, m-calpain could cleave in vitro-translated γc in a dose-dependent manner. Second, calcium could activate cleavage of γc in YT lysates, and this cleavage was diminished by calpain inhibitors. Third, ionomycin treatment of intact YT cells induced cleavage of γc in a calcium-dependent manner. Fourth, in murine thymocytes, anti-CD3 induced cleavage of γc, and this cleavage was diminished by a calpain inhibitor. Fifth, calpain inhibitors can enhance anti-CD3-induced proliferation and this increase is prevented by anti-γc blocking antibodies. Finally, calpain inhibitors can reverse ionomycin-induced decreases in STAT protein activation.
Although calpain has been implicated as playing an important role in T cell biology (24–26), the relevant substrates have remained obscure. We now show that γc is a substrate for calpain. γc is the first member of the cytokine receptor superfamily that has been shown to be a target for calpain. Although the cytoplasmic domains of IL-2Rα and IL-2Rβ are not rich in P, E, S, and T residues, a number of other cytokine receptors, including human βc (a shared component of the receptors for IL-3, IL-5, and granulocyte/macrophage colony-stimulating factor), gp130 (a shared component of the receptors for IL-6, IL-11, leukemia inhibitor factor, ciliary neurotropic factor, oncostatin M, and cardiotropin 1), IL-4Rα, IL-7Rα, and IL-9Rα have cytoplasmic domains with significant pest scores (Table 2). It will be interesting to determine if any of these cytokine receptors are also substrates for calpain.
Given that relatively high concentrations of calcium are required for activation of calpain in vitro and that antigenic peptide concentration correlates with the increase in calcium achieved (53), the strongest antigenic peptide signals would be most expected to activate calpain and promote the cleavage of γc. As γc is required for signaling in response to a variety of cytokines, the cleavage of γc could represent a mechanism by which these responses can be controlled. Because activation of mature T cells by antigen generally results in an IL-2-dependent expansion of cells, it is clear that in this context that calpain is not being activated to a sufficient extent to prevent IL-2 signaling. Thus, γc is not being quantitatively inactivated or it is being replaced at a rate sufficient to allow for γc-dependent proliferation; however, the ability of a calpain inhibitor to augment anti-CD3-induced proliferation suggests that cleavage of γc by calpain can modulate proliferation (Fig. 5).
It is interesting that TCR-induced apoptosis during intrathymic development is known to require an increase in intracellular calcium (29, 43, 45, 46) and that the strongest antigenic signals are most effective in inducing thymic or peripheral deletion of cells (54–62). It thus is conceivable that TCR-activated proteolysis of γc could predispose to apoptotic death. This type of mechanism is consistent with the ability of cytokines that signal via γc, such as IL-2 and IL-7, to protect against apoptosis in lymphocytes (48, 50, 63, 64), and suggests that inactivation of γc would diminish cytokine-induced survival signals. Nevertheless, it is clear that other mechanisms also must be operational given the very modest effect of calpain inhibitors on the ability of IL-7 to protect against anti-CD3-mediated death. In contrast, calpain inhibitors had much more potent effects related to proliferation and STAT protein activation.
Although further work is needed to clarify the physiological role that calpain plays in regulating signaling by γc-dependent cytokines, our data provide a key insight that calpain can associate with and cleave γc, suggesting that TCR-mediated proteolysis of γc by calpain could represent a mechanism by which cytokine responsiveness can be controlled, and thereby providing the basis for additional investigation.
Table 1.
TCR-induced death of thymocytes is diminished by a combination of IL-7 + calpastatin
| Calpastatin | IL-7 | % dead cells
|
% induced cell death | |
|---|---|---|---|---|
| Control | TCR-induced | |||
| − | − | 25.5 ± 0.7 | 45.7 ± 2.5 | 20.2 ± 2.6 |
| − | + | 23.0 ± 0.9 | 42.0 ± 1.7 | 19.0 ± 1.9 |
| + | − | 24.9 ± 0.3 | 45.4 ± 1.3* | 20.6 ± 1.3 |
| + | + | 23.8 ± 2.1 | 37.8 ± 1.4* | 14.0 ± 2.5 |
Thymocytes from 4-week-old C57BL/6 mice were treated with hamster IgG (control) or anti-CD3 + anti-CD28 (TCR-induced) in the presence or absence of IL-7 and calpastatin. TCR-induced cells treated with IL-7 + calpastatin had significantly less death (37.8%) than cells treated with calpastatin alone (45.5%) (∗ P < 0.05 by Student t test). The % induced cell death is the difference between the control and TCR-induced death.
Table 2.
Cytokine receptor subunits with PEST sequences in their cytoplasmic domains
| Subunits | PEST score | Position | PEST sequences* |
|---|---|---|---|
| Human γc | −3.95 | 294–305 | GLAESLQPDYSE |
| Canine γc | −3.95 | 293–304 | GLAESLQPDYSE |
| Murine γc | +0.59 | 294–305 | GLTESLQPDYSE |
| Human βc | +6.55 | 232–245 | PSPDAGEEECSPVL |
| +14.98 | 497–529 | VCDPPSGPDTTPAASDLPTEQPPSPQPGPPAAS | |
| Human gp 130 | +8.72 | 233–258 | DASTWSQIPPEDTAST |
| +0.98 | 580–589 | DGPEFTFTTP | |
| +8.05 | 717–741 | PSISSSDENESSQNTSSTVQYSTVV | |
| +10.78 | 758–769 | SESTQPLLDSEE | |
| −4.19 | 866–880 | FETVGMEAATDEGMP | |
| Human IL-4Rα | −3.92 | 381–416 | TESLFLDLLGEENGGFCQQDMGESCLLPPSGSTSA |
| +4.08 | 442–472 | LEPSPPASPTQSPDNLTCTEPLVIAGNPAY | |
| +8.22 | 498–529 | LEEVEPEMPCVPQLSEPTTVPEPETWEQIL | |
| +0.23 | 736–762 | APDPSPGGVPLEASLCPASLAPSGISE | |
| Human IL-7Rα | +4.06 | 298–316 | DEVEGLQDTFPQQLEESE |
| −1.84 | 320–340 | LGGDVQSPNCPSEDVVVTPESFG | |
| Human IL-9Rα | +3.27 | 329–341 | SVALEEEQEGPGT |
| Human IL-2Rα | No PEST sequence found | ||
| Human IL-2Rβ | No PEST sequence found | ||
Sequences were analyzed using pest-find.
Acknowledgments
We thank S. Elledge for pAS-CYH2, Y187 and Y190 yeast strains, and the Epstein–Barr virus B cell cDNA library in pACT; J. Chiorini for help with computer analysis of PEST sequences; M. Rechsteiner for the pest-find computer program; J. Yodoi for YT cells; K. Furuke for isolating peripheral blood lymphocytes; M. Martin, T. Kitamura, T. Hara, and A. Yamauchi for technical recommendations; and S. M. Russell, J. X. Lin, B. J. Fowlkes, P. E. Love, R. H. Schwartz, R. N. Germain, L. E. Samelson, and L. D’Adamio for valuable discussions and/or critical comments. M. J. A. was supported in part by Grant AM 104/1-1 from the Deutsche Forschungsgemeinschaft, Germany.
ABBREVIATIONS
- γc
common cytokine receptor γ chain
- PEST
proline, glutamate, serine, and threonine
- TCR
T cell receptor
- IL
interleukin
- β-gal
β-galactosidase
- FBS
fetal bovine serum
References
- 1.Noguchi M, Yi H, Rosenblatt H M, Filipovich A H, Adelstein S, Modi W S, McBride O W, Leonard W J. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 2.Leonard W J. Annu Rev Med. 1996;47:229–239. doi: 10.1146/annurev.med.47.1.229. [DOI] [PubMed] [Google Scholar]
- 3.Takeshita T, Asao H, Ohtani K, Ishii N, Kumaki S, Tanaka N, Munakata H, Nakamura M, Sugamura K. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 4.Kondo M, Takeshita T, Ishii N, Nakamura M, Watanabe S, Arai K, Sugamura K. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 5.Noguchi M, Nakamura Y, Russell S M, Ziegler S F, Tsang M, Cao X, Leonard W J. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 6.Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R K, Paul W E, Leonard W J. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 7.Kondo M, Takeshita T, Higuchi M, Nakamura M, Sudo T, Nishikawa S, Sugamura K. Science. 1994;263:1453–1454. doi: 10.1126/science.8128231. [DOI] [PubMed] [Google Scholar]
- 8.Russell S M, Johnston J A, Noguchi M, Kawamura M, Bacon C M, Friedmann M, Berg M, McVicar D W, Witthuhn B A, Silvennoinen O, Goldman A S, Schmalsteig F C, Ihle J N, O’Shea J J, Leonard W J. Science. 1994;266:1042–1045. doi: 10.1126/science.7973658. [DOI] [PubMed] [Google Scholar]
- 9.Giri J G, Ahdieh M, Eisenman J, Shanebeck K, Grabstein K, Kumaki S, Namen A, Park L S, Cosman D, Anderson D. EMBO J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Disanto J, Muller W, Guy-Grand D, Fischer A, Rajewsky K. Proc Natl Acad Sci USA. 1995;92:377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X, Shores E W, Hu-Li J, Anver M, Kelsall B L, Russell S M, Drago J, Noguchi M, Grinberg A, Bloom E T, Paul W E, Katz S I, Paul E L, Leonard W J. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 12.Leonard W J, Shores E W, Love P E. Immunol Rev. 1995;148:97–114. doi: 10.1111/j.1600-065x.1995.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 13.Cao X, Kozak C A, Liu Y J, Noguchi M, O’Connell E, Leonard W J. Proc Natl Acad Sci USA. 1993;90:8464–8468. doi: 10.1073/pnas.90.18.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boussiotis V A, Barber D L, Nakarai T, Freeman G J, Gribben J G, Bernstein G M, d’Andrea A D, Ritz J, Nadler L M. Science. 1994;266:1039–1042. doi: 10.1126/science.7973657. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura Y, Russell S M, Mess S A, Friedmann M, Erdos M, Francois C, Jacques Y, Adelstein S, Leonard W J. Nature (London) 1994;369:330–333. doi: 10.1038/369330a0. [DOI] [PubMed] [Google Scholar]
- 16.Nelson B, Lord J D, Greenberg P D. Nature (London) 1994;369:333–336. doi: 10.1038/369333a0. [DOI] [PubMed] [Google Scholar]
- 17.Hatakeyama M, Kono T, Kobayashi N, Kawahara A, Levin S D, Perlmutter R M, Taniguchi T. Science. 1991;252:1523–1528. doi: 10.1126/science.2047859. [DOI] [PubMed] [Google Scholar]
- 18.Ravichandran K S, Burakoff S J. J Biol Chem. 1994;269:1599–1602. [PubMed] [Google Scholar]
- 19.Friedmann M C, Migone T-S, Russell S M, Leonard W J. Proc Natl Acad Sci USA. 1996;93:2077–2082. doi: 10.1073/pnas.93.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minami Y, Nakagawa Y, Kawahara A, Miyazaki T, Sada K, Yamamura H, Taniguchi T. Immunity. 1995;2:89–100. doi: 10.1016/1074-7613(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 21.Truitt K E, Mills G E, Turck C W, Imboden J B. J Biol Chem. 1994;269:5937–5943. [PubMed] [Google Scholar]
- 22.Miyazaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Liu Z-J, Oishi I, Silvennoinen O, Witthuhn B A, Ihle J N. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 23.Lin J-X, Migone T S, Tsang M, Friedmann M, Weatherbee J A, Zhou L, Yamauchi A, Bloom E T, Mietz J, John S, Leonard W J. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 24.Squier M K T, Miller A C K, Malkinson A M, Cohen J J. J Cell Physiol. 1994;159:229–237. doi: 10.1002/jcp.1041590206. [DOI] [PubMed] [Google Scholar]
- 25.Sarin A, Nakajima H, Henkart P A. J Immunol. 1995;154:5806–5812. [PubMed] [Google Scholar]
- 26.Martin S J, Green D R. Cell. 1995;82:349–352. doi: 10.1016/0092-8674(95)90422-0. [DOI] [PubMed] [Google Scholar]
- 27.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 28.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 29.Finkel T H, Marrack P, Kappler J W, Kubo R T, Cambier J C. J Immunol. 1989;142:3006–3012. [PubMed] [Google Scholar]
- 30.Punt J A, Osborne B A, Takahama Y, Sharrow S O, Singer A. J Exp Med. 1994;179:709–713. doi: 10.1084/jem.179.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyons A B, Samuel K, Sanderson A, Maddy A H. Cytometry. 1992;13:809–821. doi: 10.1002/cyto.990130803. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki H, Emori Y, Imajoh-Ohmi S, Minami Y, Suzuki K. J Biochem. 1989;106:274–281. doi: 10.1093/oxfordjournals.jbchem.a122844. [DOI] [PubMed] [Google Scholar]
- 33.Fields S, Song O K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 34.Murachi T. Biochem Int. 1989;18:263–294. [PubMed] [Google Scholar]
- 35.Croall D E, Demartino G N. Physiol Rev. 1991;71:813–847. doi: 10.1152/physrev.1991.71.3.813. [DOI] [PubMed] [Google Scholar]
- 36.Sorimachi H, Saido T C, Suzuki K. FEBS Lett. 1994;343:1–5. doi: 10.1016/0014-5793(94)80595-4. [DOI] [PubMed] [Google Scholar]
- 37.Deshpande R V, Goust J-M, Chakrabarti A K, Barbosa E, Hogan E L, Banik N L. J Biol Chem. 1995;270:2497–2505. doi: 10.1074/jbc.270.6.2497. [DOI] [PubMed] [Google Scholar]
- 38.Rechsteiner M. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- 39.Rechsteiner M. Cell Biol. 1990;1:433–440. [PubMed] [Google Scholar]
- 40.Molinari M, Anagli J, Carafoli E. J Biol Chem. 1995;270:2032–2035. doi: 10.1074/jbc.270.5.2032. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe N, Van de Woude G, Ikawa Y, Sagata N. Nature (London) 1989;342:505–511. doi: 10.1038/342505a0. [DOI] [PubMed] [Google Scholar]
- 42.Mills G B, Cheung R K, Grinstein S, Gelfand E W. J Immunol. 1985;134:2431–2435. [PubMed] [Google Scholar]
- 43.McConkey D J, Nicotera P, Orrenius S. Immunol Rev. 1994;142:343–363. doi: 10.1111/j.1600-065x.1994.tb00896.x. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama T, Ueda Y, Yamada H, Shores E W, Singer A, June C H. Science. 1992;257:96–99. doi: 10.1126/science.1621102. [DOI] [PubMed] [Google Scholar]
- 45.Robey E, Fowlkes B J. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 46.McConkey D J, Hartzell P, Amador-Prez J F, Orrenius S, Jondal M. J Immunol. 1989;143:1801–1806. [PubMed] [Google Scholar]
- 47.Smith C A, Williams G T, Kingston R, Jenkinson E J, Owen J J T. Nature (London) 1989;337:181–184. doi: 10.1038/337181a0. [DOI] [PubMed] [Google Scholar]
- 48.Nieto M A, Gonzalez A, Lopez-Rivas A, Diaz-Espada F, Gambon F. J Immunol. 1990;145:1364–1368. [PubMed] [Google Scholar]
- 49.Otani H, Erdos M, Leonard W J. J Biol Chem. 1993;268:22733–22736. [PubMed] [Google Scholar]
- 50.Groux H, Monte D, Plouvier B, Capron A, Ameisen J-C. Eur J Immunol. 1993;23:1623–1629. doi: 10.1002/eji.1830230734. [DOI] [PubMed] [Google Scholar]
- 51.Boise L H, Minn A J, June C H, Lindsten T, Thompson C B. Proc Natl Acad Sci USA. 1995;92:5491–5495. doi: 10.1073/pnas.92.12.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Y W, Adkins B, Furse R K, Malek T R. J Immunol. 1995;154:1596–1605. [PubMed] [Google Scholar]
- 53.Vasquez N J, Kane L P, Hedrick S M. Immunity. 1994;1:45–56. doi: 10.1016/1074-7613(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 54.Sprent J, Lo D, Gao E-K, Ron Y. Immunol Rev. 1988;101:173–190. doi: 10.1111/j.1600-065x.1988.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 55.Yagi J, Janeway C A. Int Immunol. 1990;2:83–89. doi: 10.1093/intimm/2.1.83. [DOI] [PubMed] [Google Scholar]
- 56.Ashton-Rickardt P G, Bandeira A, Delaney J R, Kaer L V, Pircher H-P, Zinkernagel R M, Tonegawa S. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 57.Hogquist K A, Jameson S C, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 58.Webb S, Morris C, Sprent J. Cell. 1990;63:1249–1256. doi: 10.1016/0092-8674(90)90420-j. [DOI] [PubMed] [Google Scholar]
- 59.Webb S, Sprent J. Immunol Rev. 1989;107:141–158. doi: 10.1111/j.1600-065x.1989.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 60.Kabelitz D, Pohl T, Pechhold K. Immunol Today. 1993;14:338–339. doi: 10.1016/0167-5699(93)90231-9. [DOI] [PubMed] [Google Scholar]
- 61.Fink P J, Fang C A, Turk G L. J Immunol. 1994;152:4270–4281. [PubMed] [Google Scholar]
- 62.Critchfield J M, Racke M K, Zuniga-Pflucker J C, Cannella B, Raine C S, Goverman J, Lenardo M J. Science. 1994;263:1139–1143. doi: 10.1126/science.7509084. [DOI] [PubMed] [Google Scholar]
- 63.Deng G, Podack E R. Proc Natl Acad Sci USA. 1993;90:2189–2193. doi: 10.1073/pnas.90.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Armant M, Delespesse G, Safati M. Immunology. 1995;85:331–337. [PMC free article] [PubMed] [Google Scholar]
- 65.Hirai S, Kawasaki H, Yaniv M, Suzuki K. FEBS Lett. 1991;287:57–61. doi: 10.1016/0014-5793(91)80015-u. [DOI] [PubMed] [Google Scholar]
- 66.Bosco M C, Espinoza-Degado I, Schwabe M, Russell S M, Leonard W J, Longo D L, Varesio L. Blood. 1994;83:3462–3467. [PubMed] [Google Scholar]
- 67.Epling-Burnette P K, Wei S, Liu J H, Pericle F, Ussery D, Russell S M, Leonard W J, Djeu J Y. Eur J Immunol. 1995;25:291–294. doi: 10.1002/eji.1830250148. [DOI] [PubMed] [Google Scholar]