Abstract
Free transition metal ions oxidize lipids and lipoproteins in vitro; however, recent evidence suggests that free metal ion-independent mechanisms are more likely in vivo. We have shown previously that human ceruloplasmin (Cp), a serum protein containing seven Cu atoms, induces low density lipoprotein oxidation in vitro and that the activity depends on the presence of a single, chelatable Cu atom. We here use biochemical and molecular approaches to determine the site responsible for Cp prooxidant activity. Experiments with the His-specific reagent diethylpyrocarbonate (DEPC) showed that one or more His residues was specifically required. Quantitative [14C]DEPC binding studies indicated the importance of a single His residue because only one was exposed upon removal of the prooxidant Cu. Plasmin digestion of [14C]DEPC-treated Cp (and N-terminal sequence analysis of the fragments) showed that the critical His was in a 17-kDa region containing four His residues in the second major sequence homology domain of Cp. A full length human Cp cDNA was modified by site-directed mutagenesis to give His-to-Ala substitutions at each of the four positions and was transfected into COS-7 cells, and low density lipoprotein oxidation was measured. The prooxidant site was localized to a region containing His426 because CpH426A almost completely lacked prooxidant activity whereas the other mutants expressed normal activity. These observations support the hypothesis that Cu bound at specific sites on protein surfaces can cause oxidative damage to macromolecules in their environment. Cp may serve as a model protein for understanding mechanisms of oxidant damage by copper-containing (or -binding) proteins such as Cu, Zn superoxide dismutase, and amyloid precursor protein.
Transition metal ions such as Cu and Fe are extremely effective promoters of oxidation reactions and can cause the oxidative modification of low density lipoprotein in vitro (1, 2). The involvement of free metal ions in oxidation processes in vivo has been questioned because most of these ions are linked to prosthetic groups or are tightly sequestered by specific binding proteins, and thus the level of free Cu (and Fe) in plasma and interstitial fluids is extremely low (3). In addition, recent mass spectroscopic analysis suggests that the oxidized lipid “fingerprint” exhibited by human atherosclerotic lesions is not consistent with free metal-catalyzed processes (4). In view of these findings, recent attention has focused on the investigation of free metal ion-independent mechanisms, such as those using peroxidases (5, 6), and on metal-binding proteins (7–9), which can promote LDL oxidation.
Studies by us (10–12) and others (13–15) show that the copper protein ceruloplasmin (Cp) exhibits potent oxidant activity toward LDL in vitro. Cp is a plasma glycoprotein containing about seven Cu atoms per molecule and ≈95% of the total plasma Cu (see refs. 16 and 17 for review). It is a 132-kDa monomer comprised almost entirely of three major domains that have 35–40% sequence homology to each other (18) and to three homologous domains in factors Va and VIIIa (19). Cp is an acute phase reactant that exhibits a 2- to 3-fold increase over the unevoked concentration of 300 μg/ml in adult plasma. After myocardial infarction and other diseases accompanied by severe inflammation, the Cp and Cu content of human plasma changes in parallel, indicating a tightly coupled process or at least coordinate regulation (20). Highly purified human Cp (21) increases LDL oxidation by up to 50-fold at subphysiological concentrations. Flame atomic absorption spectroscopy shows that Chelex 100 (but not extensive dialysis) removes one of the seven Cu atoms and totally inhibits the prooxidant activity, revealing the critical role of a single Cu (12). Inactivation by Chelex 100 is not due to gross or irreversible alterations in protein structure because the distinct ferroxidase activity of Cp is not altered, and prooxidant activity of Chelex 100-treated Cp is completely restored upon repletion with CuSO4. The function of Cp prooxidant activity is not known, but it may be involved in host defense; a reported bactericidal activity indicates that Cp may participate in the inflammatory response to foreign agents (22). Inflammatory cytokines induce production of Cp by monocytic cells (23, 24).
The removal of Cp prooxidant Cu by a solid-phase chelator suggests that the atom may be located at or near the surface of the protein. Oxidative damage by Cu exposed at protein surfaces has been proposed as a cause of tissue damage by certain Cu, Zn superoxide dismutase (SOD1) mutants in familial amyotrophic lateral sclerosis (25–27), and amyloid precursor protein in Alzheimer disease (28). In contrast, Cu sequestration by binding proteins may afford protection from Cu-induced oxidant injury. The high susceptibility to Alzheimer (and cardiovascular) disease in populations with a high prevalence of the E4 allele of apolipoprotein E (29) may be due to the weak Cu binding and low antioxidant activity of E4 (compared with other isoforms) (30).
Little is known about the specific environment of a protein surface that permits Cu to bind and exhibit prooxidant activity. In this report we use biochemical and molecular approaches to show that the Chelex-removable, prooxidant Cu is bound to Cp via a single, specific His residue, in a site located in a narrow valley within the second major sequence homology domain. Cp presents an effective model system for molecular studies of the mechanisms of oxidative damage by Cu atoms bound to protein surfaces and of the specific protein environment necessary for this activity.
MATERIALS AND METHODS
Reagents.
Plasmin and intact, purified human Cp were obtained from Calbiochem. Cp purity was >95% as shown by Coomassie-stained SDS/PAGE gels and was verified by an absorbance ratio (610 nm/280 nm) higher than 0.045. Densitometric analysis of immunoblotted SDS/PAGE gels showed that ≈80–85% was present as the intact, oxidant 132-kDa form of Cp. Chelex 100 was from Bio-Rad. Diethylpyrocarbonate (DEPC), [14C]DEPC (2.6 mCi/mmol), hydroxylamine, and other assay reagents were from Sigma. Human LDL (density = 1.019–1.063 g/ml) was prepared from freshly drawn, citrated, normolipemic plasma by sequential ultracentrifugation. PCR was done using oligonucleotides from Operon Technologies (Alameda, CA), and Pfu polymerase was from Stratagene.
Construction of Cp Expression Vector, Mutagenesis, and Cell Transfection.
A full length human Cp cDNA engineered to contain a consensus Kozak sequence for translation (generously provided by Jonathan Gitlin, Washington University) was subcloned into the HindIII site behind the cytomegalovirus promoter of the mammalian expression vector pcDNA3/Amp (Invitrogen). The integrity and orientation of the insert was verified by restriction mapping and sequencing. Mutagenesis of the Cp coding region was done using the PCR-based megaprimer method (31) and was verified by DNA sequencing. Plasmid DNA was transiently transfected into COS-7 cells using lipofectamine in serum-free OptiMEM medium. The medium was replaced with fresh serum-free medium and collected after 48 h.
Assay Methods.
Lipoprotein oxidation was measured as formation of thiobarbituric acid-reacting substances by a modification of the method of Schuh et al. (32). Ferroxidase activity of Cp was measured by an “in-gel” assay in which Cp-catalyzed oxidation of ferrous ion, in the presence of acidified potassium ferrocyanide, results in the formation of Prussian blue (24, 33). To measure LDL oxidation, cell-conditioned medium was concentrated by ultrafiltration using Centricon-30 filters (Amicon, Beverly, MA) and washing with PBS. The concentrate was incubated with LDL (0.5 mg cholesterol/ml), xanthine (1 mM), and xanthine oxidase (0.15 munits), and oxidation was measured after 24 h as thiobarbituric acid-reacting substances. Cp in the conditioned medium was determined by immunoblot analysis using rabbit anti-human Cp IgG (Accurate Chemicals) and visualization by chemiluminescence (enhanced chemiluminescence, Amersham).
RESULTS
The Role of His Residues in Cp Prooxidant Activity.
Cu is often bound to proteins via coordination with one or more His imidazole rings. We thus examined the ability of the His-specific modifier DEPC (34) to block the binding of the prooxidant Cu to Cp using a strategy involving Cu removal, DEPC treatment, and finally Cu repletion. Native Cp increased LDL oxidation by more than 10-fold (Table 1). Pretreatment of Cp with Chelex 100, under conditions that remove a single Cu atom (12, 35), blocked oxidation. The activity was completely restored upon subsequent incubation with CuSO4 (followed by dialysis to remove unbound Cu) as described (12). In contrast, when Chelex 100-inactivated Cp was pretreated with DEPC, subsequent incubation with CuSO4 did not restore prooxidant activity, indicating the importance of one or more His residues. Controls showed that the inactivation by DEPC was not due to nonspecific inactivation of Cp because treatment of native Cp with DEPC did not inhibit prooxidant activity. Because the specificity of DEPC for His is not absolute [it has limited reactivity toward Cys, Lys, and Tyr (36)], the spectral absorbance shift and reversibility by hydroxylamine (36) were examined, and the results were found to be consistent with specific His modification (not shown).
The number of His residues involved in the binding of the prooxidant Cu was quantitated by measuring the amount of DEPC bound to native and Chelex 100-treated Cp by the absorbance change at 244 nm and by the binding of [14C]DEPC. Both methods showed that ≈5 of 41 total His residues in native Cp are modified by DEPC under these conditions (Table 2). These residues are likely to reflect those at or near the protein surface, not coordinated to Cu, and accessible to DEPC. Prior removal of the prooxidant Cu by Chelex 100 increased the number of DEPC-modifiable residues by ≈0.7–0.8 nmol/nmol of Cp. When Chelex 100-treated Cp was repleted with Cu and then DEPC binding was measured, the data (from both methods) suggest that ≈1.1 additional His residues were exposed in Chelex 100-treated Cp. These data are consistent with a model in which a single DEPC-modifiable His residue was exposed by removal of the prooxidant Cu.
Biochemical Localization of the Prooxidant Cu Domain.
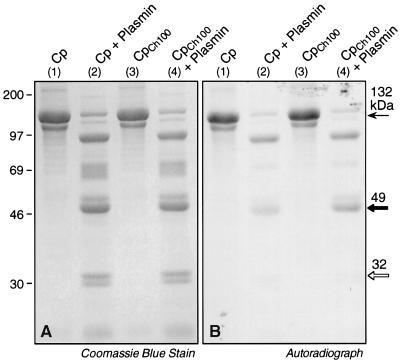
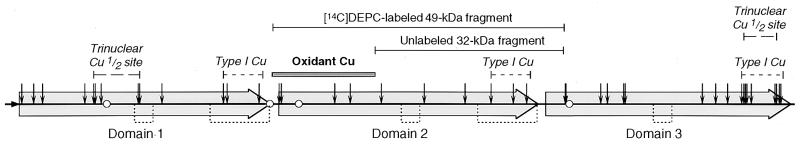
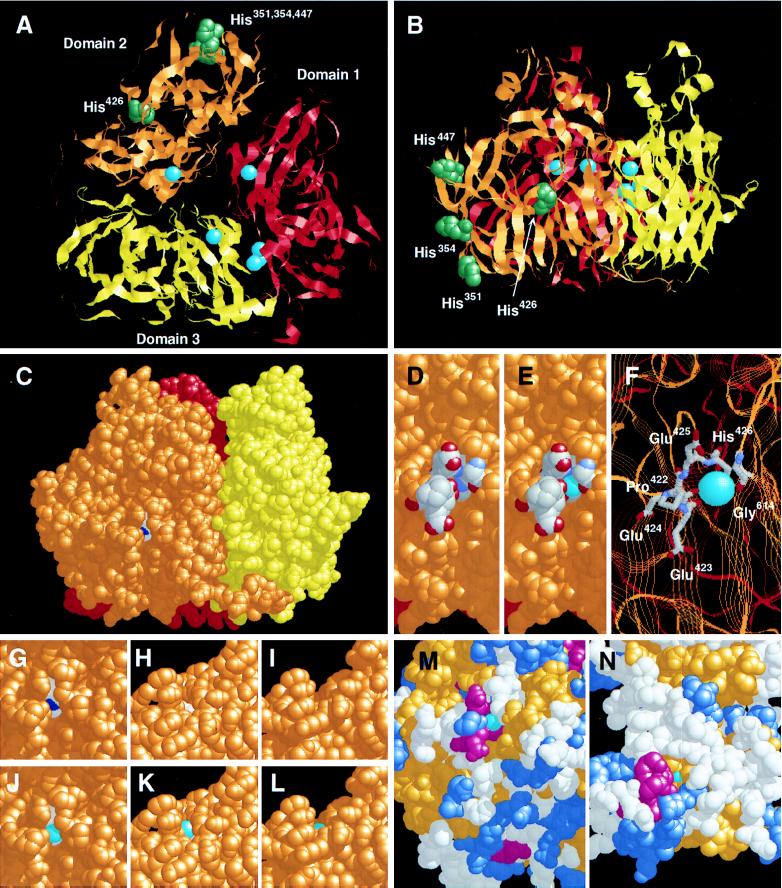
Limited proteolysis of [14C]DEPC-labeled Cp was used to determine the location of the prooxidant Cu site in Cp. Native Cp was pretreated with unlabeled DEPC to block exposed His sites not involved in prooxidant Cu binding. The prooxidant Cu was subsequently removed with Chelex 100, and newly exposed His was reacted with [14C]DEPC. The labeled protein was cleaved by limited plasmin digestion and subjected to SDS/PAGE. Coomassie blue staining showed identical fragments in native Cp (Fig. 1A, lane 2) and in Cp pretreated with Chelex 100 (Fig. 1A, lane 4). Autoradiography showed [14C]DEPC labeling primarily in two fragments of ≈90 and 49 kDa (Fig. 1B). Only the 49-kDa fragment showed higher labeling after removal of the prooxidant Cu (lane 4 vs. lane 2). N-terminal analysis gave a sequence consistent with S341SSKDNIRGKHVRHYYIAA, a region near the N terminus of the second major sequence homology domain [Fig. 2; we use the nomenclature in which Cp is comprised of three internally triplicated domains or “homology units” (18)]. An unlabeled 32-kDa fragment gave a sequence consistent with S482VPPSASHVAPTETFTYEWT. By the difference between the labeled and unlabeled fragments, the critical His residue was determined to be in a 17-kDa region containing four His residues near the N terminus of domain 2. The x-ray structure of Cp (37) shows that three of the His residues in this region, His351, His354, and His447, are at the surface and that His426 is in a shallow valley near the surface (Fig. 3 A and B).
Figure 1.
Identification of prooxidant site by limited plasmin digestion of [14C]DEPC-treated Cp. Purified human Cp (100 μg) was pretreated with unlabeled DEPC (1 mM) to block uninvolved His. After washing to remove unreacted DEPC, aliquots were incubated with Chelex 100. Native and Chelex 100-treated Cp were incubated with [14C]DEPC for 2 h as described in Table 1. Unreacted [14C]DEPC was removed by ultrafiltration, and Cp was incubated with plasmin (plasmin-to-Cp ratio 1:50, wt/wt) for 1 h at 23°C. Samples containing 50 μg of Cp were added to SDS sample buffer, boiled for 3 min, and subjected to electrophoresis. (A) Gel stained with Coomassie blue. Native Cp (lane 1); native Cp digested with plasmin (lane 2); Chelex 100-treated Cp (CpCh100, lane 3); CpCh100 digested with plasmin (lane 4). (B) The stained gel was transferred to Immobilon-P (Millipore) membranes and subjected to fluorography. The lanes are labeled as in A. At right, intact Cp is indicated by a thin arrow, the labeled 49-kDa fragment is indicated by a solid arrow, and the unlabeled 32-kDa fragment is indicated by an open arrow. The position of molecular weight standards is shown at left.
Figure 2.
Linear map of prooxidant domain of human Cp. Shown are the [14C]DEPC-labeled 49-kDa and unlabeled 29-kDa fragments (solid bars) and the derived prooxidant Cu site (filled bar). Also shown are the signal sequence (dark arrow), major sequence homology domains (broad gray arrows), N-linked glycosylation sites (circles), disulfide bonds (dotted lines), His residues (vertical arrows), type 1 Cu sites (dotted bars), and trinuclear Cu sites (dashed bars).
Figure 3.
Structure of the Cp prooxidant site. RasMol was used for modeling Cp using x-ray crystallographic coordinates at 3.1 Å resolution. (A) “Ribbon” diagram showing axial (“top”) view of the three major sequence homology domains in red (domain 1), orange (domain 2), and yellow (domain 3). Endogenous Cu atoms are shown as cyan spheres. His residues subjected to site-directed mutagenesis are shown in green. (B) Same as A but in side-view. (C) Spacefill drawing (with shadowing to emphasize depth) showing His426 in white and the imidazole-N in dark blue. Domain colors are as in (A). (D) Close view of prooxidant site; surface amino acids nearest to His426 are shown in CPK colors (gray = C, blue = N, red = O). (E) Same as D, but putative location of prooxidant Cu is shown as cyan sphere. (F) Enlarged stick diagram of prooxidant site showing predicted nearest amino acids. (G–I) Close view of prooxidant site as in C and rotation (with top rotating away from viewer) to more clearly illustrate the narrow valley. (J–L) Same as G–I but with prooxidant Cu in position. (M) Secondary structure model of prooxidant domain of Cp showing residues participating in α-helices as magenta, β-sheets as gold, turns as medium blue, Cu as cyan, and other residues as white. (N) Secondary structure model of Cu site in SOD1; color key is the same as in M.
Fine-Mapping of the Prooxidant Domain of Cp by Mutagenesis.
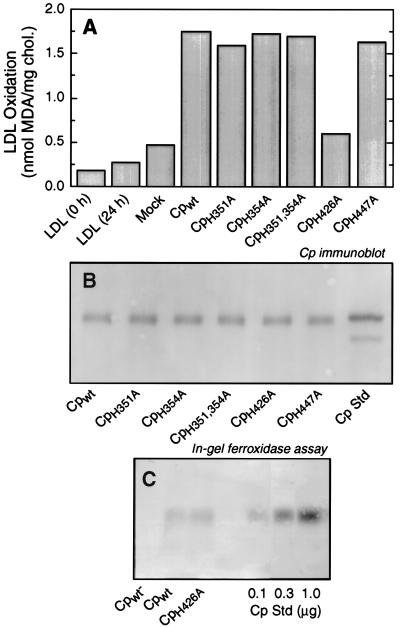
To determine the specific His required for prooxidant activity, we used a mammalian cDNA expression system and site-directed mutagenesis. The megaprimer method (31) was used to modify the full length human Cp cDNA in pcDNA3/Amp (Invitrogen) so that each of the four candidate His residues was mutated to Ala in the protein product. The two adjacent residues, His351 and His354, were also mutated in tandem. Plasmids containing wild-type and mutated Cp cDNA were transiently transfected into COS-7 cells using lipofectamine, and the conditioned medium was collected after 48 h. The medium was concentrated and added to LDL in the presence of xanthine and xanthine oxidase [an exogenous superoxide-generating system added to overcome the nonspecific inhibitory activity of secreted proteins (11)], and LDL oxidation was measured as thiobarbituric acid-reacting substances. The wild-type Cp and Cp-containing His-to-Ala mutations in positions 351, 354, 351, and 354 in tandem and 447 all had high and nearly equivalent prooxidant activity (Fig. 4A). In contrast, Cp with a His-to-Ala mutation in position 426 had only slightly more than the mock-transfected control. Immunoblot analysis of conditioned medium using rabbit anti-human Cp IgG showed that the wild-type and all mutant Cp species were secreted to about the same extent (Fig. 4B). To assess the possibility that the His426 mutation caused a global disruption that inactivated the protein, a sensitive in-gel assay was used to measure the independent Cp ferroxidase activity. This activity of Cp is thought to require at least four and possibly all six internal Cu atoms (37), and we have shown previously that this oxidase activity of Cp is independent of the presence of the prooxidant Cu (12). Conditioned medium from COS-7 cells transfected with plasmids containing Cpwild-type and CpH426A cDNA exhibited nearly identical ferroxidase activity, showing that the absence of prooxidant activity was not due to a nonspecific global effect of the CpH426A mutant (Fig. 4C).
Figure 4.
Prooxidant activity of recombinant Cp containing His-to-Ala mutations. COS-7 cells in 35-mm dishes were transfected with cDNAs (2 μg) containing wild-type (Cpwt) and mutant Cp using lipofectamine. After 48 h, the conditioned medium was collected and concentrated by ultrafiltration. (A) LDL oxidation by concentrated conditioned medium in the presence of xanthine/xanthine oxidase was measured after 24 h as thiobarbituric acid-reacting substances. (B) Synthesis and release of Cp from COS-7 cells transfected with wild-type and mutant cDNAs were measured by immunoblot analysis of 50 μl of conditioned medium using rabbit anti-human Cp IgG and visualization by chemiluminescence. A standard (Cp Std) containing 30 ng of Cp purified from human serum is included. (C) Ferroxidase activity. Conditioned media from COS-7 cells transfected with cDNAs for Cp in the reverse orientation (Cpwt−), wild-type Cp (Cpwt) and the CpH426A mutant, and Cp purified from human serum were subjected to PAGE under nondenaturing conditions and ferroxidase activity measured by an in-gel assay.
DISCUSSION
Most previous information on Cp structure and function has been derived from analogy to homologous copper-containing proteins. The results described here are the first to use cDNA mutagenesis and expression to directly probe the specific domain responsible for any Cp function. In addition, the recent elucidation of the x-ray structure of Cp by one of the authors (37) now permits functional analysis to be understood in the context of specific three-dimensional structures. The location of His426 in the second domain is shown in a shadowed, space-fill model of Cp (Fig. 3C). Our data suggest that His426 is a residue critical for LDL oxidation by Cp and are consistent with a model in which His426 is part of the binding site of the prooxidant Cu. We have modeled the putative binding site by placing the prooxidant Cu atom next to the His426 imidazole-N by equalizing distances between the Cu and all of its nearest neighbors (Fig. 3 D and E). A “stick-figure” identifies these neighbors as Pro422, Glu423, His426, and Gly614 (in E419RGPEEEHLGILGP… . G614LTM, the nearest amino acid neighbors are italicized) and that the nearest atoms are all electronegative (Fig. 3F). The prooxidant Cu site is near the bottom of a narrow “valley,” which is illustrated by a shadowing function of the molecular model (Fig. 3G) and more clearly by rotation of the model (top rotating away from the viewer) to a “side-view” (Fig. 3 G–I). Rotation of the molecular model with the prooxidant Cu in place is shown in Fig. 3 J–L.
Interest in proteins with Cu, or Cu-binding sites, at their surface has increased during the last several years, particularly in the areas of human vascular disease and neuropathology. Of the known proteins with a surface-exposed Cu atom, perhaps the best understood at the functional level is SOD1. The Cu site of SOD1 (Fig. 3N) appears to be only slightly less exposed than that of the prooxidant Cu in Cp (Fig. 3M), but both Cu atoms are associated with short, isolated α-helical stretches forming one valley “wall.” Mutations in SOD1 are responsible for a clinically important form of familial amyotrophic lateral sclerosis. Recent studies suggest that not all familial amyotrophic lateral sclerosis-linked SOD1 mutations cause loss of dismutase activity. Some SOD1 mutations are thought to cause neuropathologic damage due to a “gain-of-function” in which unnatural substrates have access to the active site and are oxidatively damaged (or initiate oxidative processes) by Cu-mediated reactions (25–27). A proposed mechanism for oxidative injury by SOD1 is that mutations near the Cu binding site increase the surface exposure of the Cu atom (25), possibly to an extent comparable to that of Cp.
There is, at present, no structural model that explains in detail the prooxidant activity of Cu bound to protein surfaces. Identification of specific prooxidant sites is an important first step toward understanding the mechanisms of oxidant damage by Cp and its potential pathophysiologic role. Many important and fundamental questions remain unanswered, e.g., the redox nature of the prooxidant Cu, the possible role of oxo-His formation or a His protonation–deprotonation cycle in the oxidation process (38), and the nature of the interaction among the prooxidant site and LDL and other substrates, to list just a few. In addition, Cp may serve as a model for understanding the structure and oxidative function of other Cu-containing proteins. Cp, with its three homologous domains containing very similar structures but with very different activities, may provide a unique experimental system for addressing basic questions of Cu–protein interactions and activities. The role of Cp oxidant activity in inflammation and other pathological processes is not known. The strong association of serum Cp and coronary heart disease (39, 40) and the presence of immunodetectable Cp in human atherosclerotic lesions (41, 42) are consistent with a role of Cp prooxidant activity in vascular disease progression. The new information on the specific prooxidant site of Cp should permit genetic approaches in which specific functions of Cp are altered in transgenic animals, providing a powerful tool for assessing the normal and pathological function(s) of Cp in vivo.
Table 1.
Effect of DEPC on Cp prooxidant activity
| LDL oxidation TBARS, nmol MDA/mg cholesterol | Inhibition, % | |
|---|---|---|
| 1. LDL only | 1.4 ± 0.2 | |
| 2. +Cp | 17.3 ± 1.1 | |
| 3. +CpCh100 | 1.6 ± 0.2 | 99 |
| 4. +CpCh100; +Cu | 16.8 ± 0.9 | 3 |
| 5. +CpCh100; +DEPC; +Cu | 1.5 ± 0.2 | 99 |
| 6. +Cp; +DEPC; | 17.8 ± 1.0 | −3 |
| 7. +CpCh100; +Cu; +DEPC | 17.1 ± 0.8 | 1 |
LDL (0.5 mg cholesterol/ml) was incubated in the absence (row 1) or presence (row 2) of Cp (100 μg/ml) for 20 h at 37°C, and oxidation was measured as TBARS. LDL was incubated with Cp subject to various treatments (rows 3–7). Cp was incubated with Chelex 100 (CpCh100, 100μl of beads washed with metal ion-free PBS/100 μg of protein) for 2 h at 23°C, followed by low speed centrifugation (row 3). CpCh100 was then incubated with 5 μM CuSO4 for 2 h to replenish bound Cu and washed extensively by ultrafiltration (row 4). CpCh100 was incubated with 150 μM DEPC for 1 h, unreacted material was removed by ultrafiltration, and Cu was replenished as above (row 5). As controls to show that DEPC did not alter the activity of Cu-full Cp, native Cp was treated with DEPC (row 6), or CpCh100 was repleted with Cu before treatment with DEPC (row 7). TBARS, thiobarbituric acid-reacting substances; MDA, malondialdehyde.
Table 2.
Quantitation of DEPC-modifiable His residues
| Cp treatment | His residues modified by DEPC nmol His/nmol Cp
|
|
|---|---|---|
| By ΔA244nm | By [14C]DEPC binding | |
| 1. Untreated | 5.0 ± 0.2 (0.7) | 5.3 ± 0.1 (0.8) |
| 2. Chelex 100 | 5.7 ± 0.2 | 6.1 ± 0.3 |
| 3. Chelex 100; Cu2+ | 4.6 ± 0.2 (1.1) | 5.0 ± 0.3 (1.1) |
Purified human Cp (row 1) was pretreated with Chelex 100 to remove the prooxidant Cu (row 2) and then with 5 μM CuSO4 to replete the Cu (row 3). The treated Cp preparations (10 μM) were incubated with 1 mM DEPC in PBS for 1 h. The number of modified His residues was determined by the increase in A244nm, using a molar extinction coefficient of 3200 M−1·cm−1. The difference compared with Chelex 100-treated Cp is shown in parentheses. Cp (5 nmol) was also incubated with 500 nmol of [14C]DEPC for 1 h at 23°C in 0.5 ml of PBS. Unbound DEPC was removed by ultrafiltration using a Microcon-10 filter (Amicon), and bound [14C]DEPC was measured by liquid scintillation counting.
Acknowledgments
This work was supported by Grants HL29582 and HL52692 from the National Heart, Lung, and Blood Institute and the National Institutes of Health (to P.L.F.) and by a Grant-in-Aid (to P.L.F.) and Fellowship (to C.K.M.) from the American Heart Association, Northeast Ohio Affiliate.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: Cp, ceruloplasmin; DEPC, diethylpyrocarbonate; LDL, low density lipoprotein; SOD1, Cu, Zn superoxide dismutase.
References
- 1.Steinberg D. Adv Exp Med Biol. 1995;369:39–48. doi: 10.1007/978-1-4615-1957-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Chisolm G M, Penn M S. In: Atherosclerosis and Coronary Artery Disease. Fuster V, Ross R, Topol E, editors; Fuster V, Ross R, Topol E, editors. New York: Raven; 1995. [Google Scholar]
- 3.Halliwell B, Gutteridge J M C. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 4.Leeuwenburgh C, Rasmussen J E, Hsu F F, Mueller D M, Pennathur S, Heinecke J W. J Biol Chem. 1997;272:3520–3526. doi: 10.1074/jbc.272.6.3520. [DOI] [PubMed] [Google Scholar]
- 5.Wieland E, Parthasarathy S, Steinberg D. Proc Natl Acad Sci USA. 1993;90:5929–5933. doi: 10.1073/pnas.90.13.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinecke J W, Li W, Francis G A, Goldstein J A. J Clin Invest. 1993;91:2866–2872. doi: 10.1172/JCI116531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdalla D S P, Campa A, Monteiro H P. Atherosclerosis. 1992;97:149–159. doi: 10.1016/0021-9150(92)90128-4. [DOI] [PubMed] [Google Scholar]
- 8.Balla G, Jacob H S, Eaton J W, Belcher J D, Vercellotti G M. Arterioscler Thromb. 1991;11:1700–1711. doi: 10.1161/01.atv.11.6.1700. [DOI] [PubMed] [Google Scholar]
- 9.Lamb D J, Leake D S. FEBS Lett. 1994;352:15–18. doi: 10.1016/0014-5793(94)00903-1. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenwald E, Fox P L. J Clin Invest. 1996;97:884–890. doi: 10.1172/JCI118491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay C K, Ehrenwald E, Fox P L. J Biol Chem. 1996;271:14773–14778. doi: 10.1074/jbc.271.25.14773. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenwald E, Chisolm G M, Fox P L. J Clin Invest. 1994;93:1493–1501. doi: 10.1172/JCI117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Lenten B J, Hama S Y, De Beer F C, Stafforini D M, McIntyre T M, Prescott S M, La Du B N, Fogelman A M, Navab M. J Clin Invest. 1995;96:2758–2767. doi: 10.1172/JCI118345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb D J, Leake D S. FEBS Lett. 1994;338:122–126. doi: 10.1016/0014-5793(94)80348-x. [DOI] [PubMed] [Google Scholar]
- 15.Swain J A, Darley-Usmar V, Gutteridge J M C. FEBS Lett. 1994;342:49–52. doi: 10.1016/0014-5793(94)80582-2. [DOI] [PubMed] [Google Scholar]
- 16.Rydén L. In: Copper Proteins and Copper Enzymes. Lontie R, editor; Lontie R, editor. Vol. 3. Boca Raton, FL: CRC; 1984. pp. 37–100. [Google Scholar]
- 17.Fox P L, Mukhopadhyay C, Ehrenwald E. Life Sci. 1995;56:1749–1758. doi: 10.1016/0024-3205(95)00146-w. [DOI] [PubMed] [Google Scholar]
- 18.Ortel T L, Takahashi N, Putnam F W. Proc Natl Acad Sci USA. 1984;81:4761–4765. doi: 10.1073/pnas.81.15.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Church W R, Jernigan R L, Toole J, Hewick R M, Knopf J, Knutson G J, Nesheim M E, Mann K G, Fass D N. Proc Natl Acad Sci USA. 1984;81:6934–6937. doi: 10.1073/pnas.81.22.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adelstein S J, Coombs T L, Vallee B L. N Engl J Med. 1956;255:105–109. doi: 10.1056/NEJM195607192550301. [DOI] [PubMed] [Google Scholar]
- 21.Ehrenwald E, Fox P L. Arch Biochem Biophys. 1994;309:392–395. doi: 10.1006/abbi.1994.1129. [DOI] [PubMed] [Google Scholar]
- 22.Klebanoff S J. Arch Biochem Biophys. 1992;295:302–308. doi: 10.1016/0003-9861(92)90522-x. [DOI] [PubMed] [Google Scholar]
- 23.Fleming R E, Whitman I P, Gitlin J D. Am J Physiol. 1991;260:L68–L74. doi: 10.1152/ajplung.1991.260.2.L68. [DOI] [PubMed] [Google Scholar]
- 24.Mazumder B, Mukhopadhyay C K, Prok A, Cathcart M K, Fox P L. J Immunol. 1997;159:1938–1944. [PubMed] [Google Scholar]
- 25.Wiedau-Pazos M, Goto J J, Rabizadeh S, Gralla E B, Roe J A, Lee M K, Valentine J S, Bredesen D E. Science. 1996;271:515–518. doi: 10.1126/science.271.5248.515. [DOI] [PubMed] [Google Scholar]
- 26.Siddique T, Deng H-X. Hum Mol Genet. 1996;5:1465–1470. doi: 10.1093/hmg/5.supplement_1.1465. [DOI] [PubMed] [Google Scholar]
- 27.Yim M B, Kang J H, Yim H S, Kwak H S, Chock P B, Stadtman E R. Proc Natl Acad Sci USA. 1996;93:5709–5714. doi: 10.1073/pnas.93.12.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Multhaup G, Schlicksupp A, Hesse L, Beher D, Ruppert T, Masters C L, Beyreuther K. Science. 1996;271:1406–1409. doi: 10.1126/science.271.5254.1406. [DOI] [PubMed] [Google Scholar]
- 29.Corder E H, Saunders A M, Strittmatter W J, Schmechel D E, Gaskell P C, Small G W, Roses A D, Haines J L, Pericak-Vance M A. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 30.Miyata M, Smith J D. Nat Genet. 1996;14:55–61. doi: 10.1038/ng0996-55. [DOI] [PubMed] [Google Scholar]
- 31.Barik S. In: Methods in Molecular Biology. White B A, editor; White B A, editor. Vol. 15. Totowa, NJ: Humana; 1993. pp. 277–286. [DOI] [PubMed] [Google Scholar]
- 32.Schuh J, Fairclough G F, Haschemeyer R H. Proc Natl Acad Sci USA. 1978;75:3173–3177. doi: 10.1073/pnas.75.7.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schen R J, Rabinovitz M. Clin Chim Acta. 1966;13:537–538. doi: 10.1016/0009-8981(66)90253-1. [DOI] [PubMed] [Google Scholar]
- 34.Dumas D P, Raushel F M. J Biol Chem. 1990;265:21498–21503. [PubMed] [Google Scholar]
- 35.Huber C T, Frieden E. J Biol Chem. 1970;245:3973–3978. [PubMed] [Google Scholar]
- 36.Miles E W. Methods Enzymol. 1977;47:431–442. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- 37.Zaitseva I, Zaitsev V, Card G, Moshkov K, Bax B, Ralph A, Lindley P. J Biol Inorg Chem. 1996;1:15–23. [Google Scholar]
- 38.Morgan J E, Verkhovsky M I, Wikstrom M. J Bioenerg Biomembr. 1994;26:599–608. doi: 10.1007/BF00831534. [DOI] [PubMed] [Google Scholar]
- 39.Mänttäri M, Manninen V, Huttunen J K, Palosuo T, Ehnholm C, Heinonen O P, Frick M H. Eur Heart J. 1994;15:1599–1603. doi: 10.1093/oxfordjournals.eurheartj.a060440. [DOI] [PubMed] [Google Scholar]
- 40.Reunanen A, Knekt P, Aaran R-K. Am J Epidemiol. 1992;136:1082–1090. doi: 10.1093/oxfordjournals.aje.a116573. [DOI] [PubMed] [Google Scholar]
- 41.Hollander W, Colombo M A, Kirkpatrick B, Paddock J. Atherosclerosis. 1979;34:391–405. doi: 10.1016/0021-9150(79)90064-9. [DOI] [PubMed] [Google Scholar]
- 42.Swain J, Gutteridge J M C. FEBS Lett. 1995;368:513–515. doi: 10.1016/0014-5793(95)00726-p. [DOI] [PubMed] [Google Scholar]