Abstract
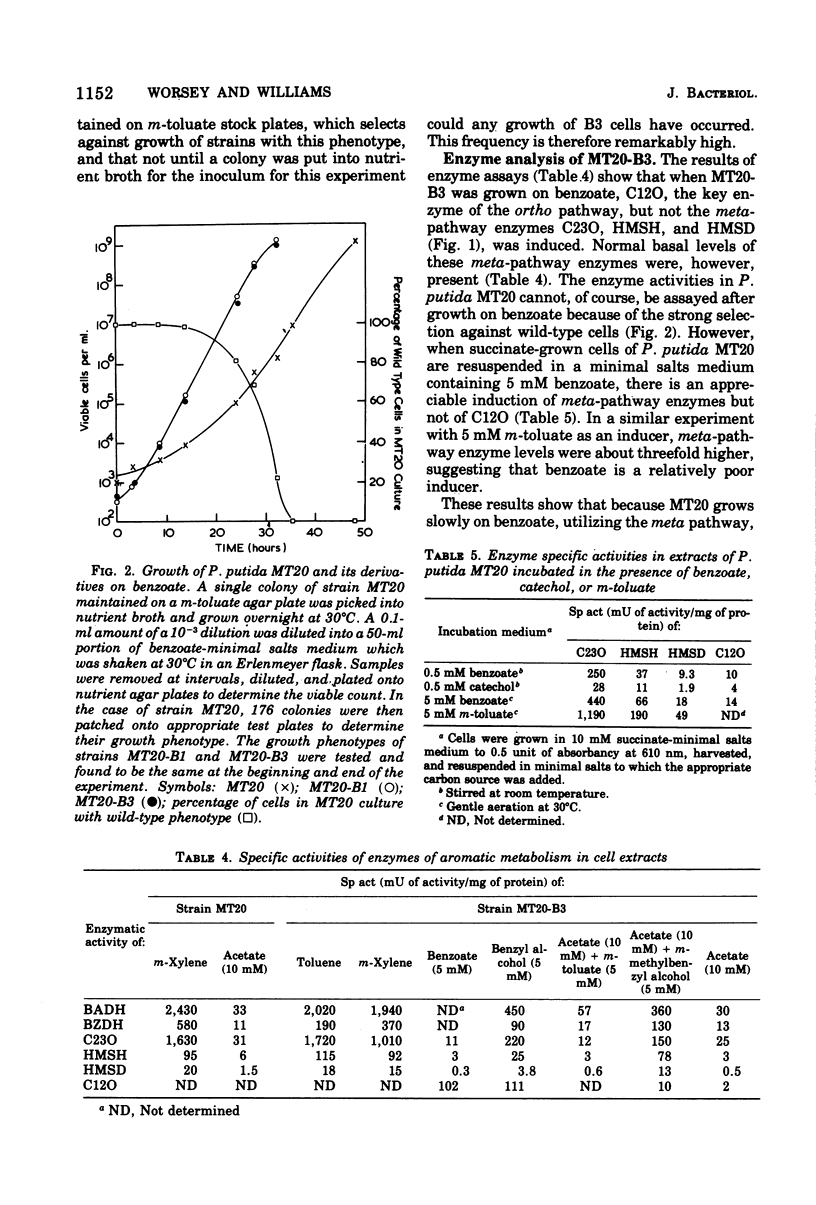
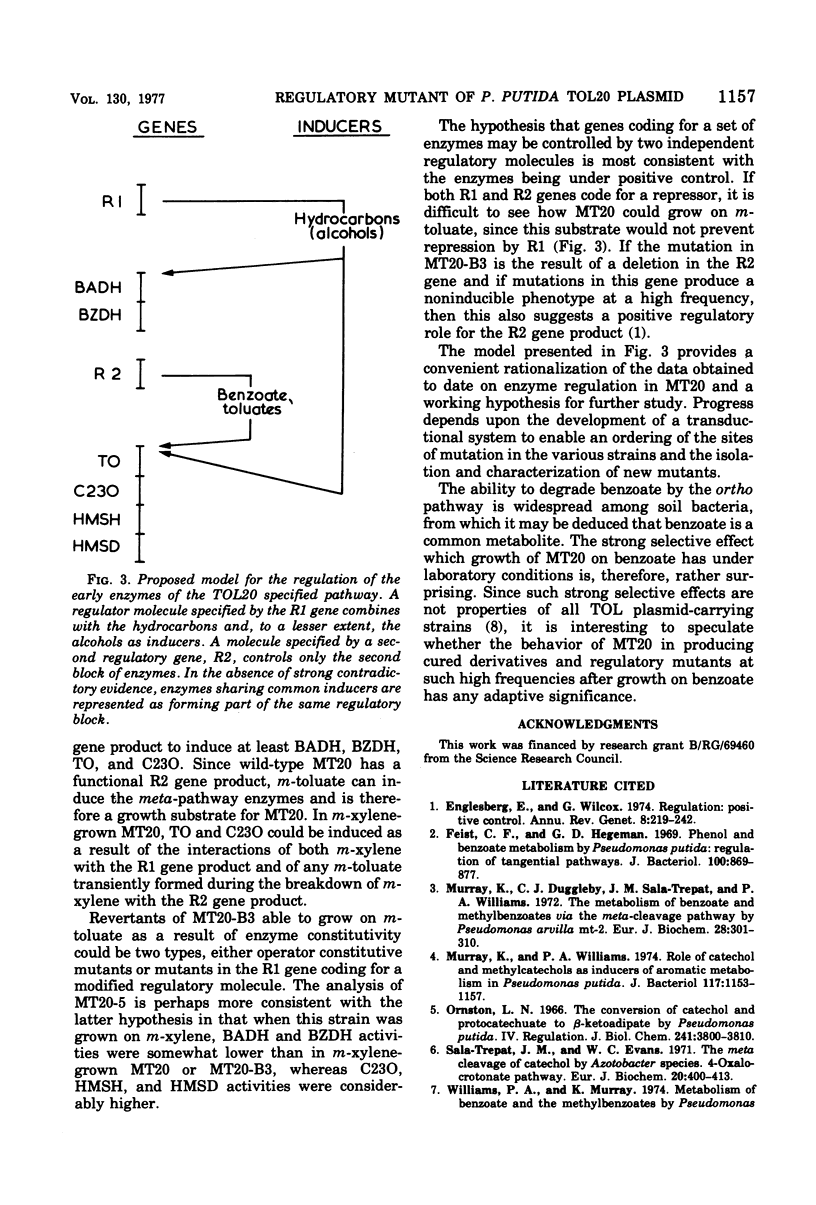
Pseudomonas putida MT20 carries a plasmid (TOL20) that codes for the enzymes responsible for the catabolism of toluene, m- and p-xylene to benzoate, and m- and p-toluate, respectively, followed by meta cleavage of the aromatic ring. Growth on 5 mM benzoate selects very strongly for (i) strains that have been cured of the plasmid and (ii) strains with an intermediate growth pattern (the B3 phenotype) that retain the ability to grow on toluene, m-xylene, and benzoate but are unable to grow on m-toluate. Both types of strains were selected because they are no longer able to oxidize benzoate by the plasmid pathway but instead use an alternative route, the ortho or β-ketoadipate pathway, which is chromosomally coded and supports faster growth. Evidence that one strain with the B3 phenotype, MT20-B3, has a regulatory mutation that prevents induction of the meta-pathway enzymes by benzoate and m-toluate, but which enables them to be induced by toluene and m-xylene, is presented. The plasmid in this strain, as in most of the others with the same phenotype, is nonconjugative. Analysis of MT20-B3, together with revertants of it and other noninducible mutants, has led to a model for the regulation of the plasmid-coded enzymes in MT20, in which it is proposed that the early enzymes for degradation of m-toluate and benzoate are positively controlled by two regulator molecules, one of which interacts with toluene and m-xylene as inducers and the other of which interacts with benzoate and m-toluate. It is argued that MT20-B3 and strains with a similar phenotype arose as a result of a deletion of the gene coding for the second regulator molecule.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Englesberg E., Wilcox G. Regulation: positive control. Annu Rev Genet. 1974;8:219–242. doi: 10.1146/annurev.ge.08.120174.001251. [DOI] [PubMed] [Google Scholar]
- Feist C. F., Hegeman G. D. Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J Bacteriol. 1969 Nov;100(2):869–877. doi: 10.1128/jb.100.2.869-877.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Duggleby C. J., Sala-Trepat J. M., Williams P. A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972 Jul 24;28(3):301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- Murray K., Williams P. A. Role of catechol and the methylcatechols as inducers of aromatic metabolism in Pseudomonas putida. J Bacteriol. 1974 Mar;117(3):1153–1157. doi: 10.1128/jb.117.3.1153-1157.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. IV. Regulation. J Biol Chem. 1966 Aug 25;241(16):3800–3810. [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Worsey M. J. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol. 1976 Mar;125(3):818–828. doi: 10.1128/jb.125.3.818-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]



