Abstract
Cancer is a disease that begins with mutation of critical genes: oncogenes and tumor suppressor genes. Our research on carcinogenic aromatic hydrocarbons indicates that depurinating hydrocarbon–DNA adducts generate oncogenic mutations found in mouse skin papillomas (Proc. Natl. Acad. Sci. USA 92:10422, 1995). These mutations arise by mis-replication of unrepaired apurinic sites derived from the loss of depurinating adducts. This relationship led us to postulate that oxidation of the carcinogenic 4-hydroxy catechol estrogens (CE) of estrone (E1) and estradiol (E2) to catechol estrogen-3,4-quinones (CE-3, 4-Q) results in electrophilic intermediates that covalently bind to DNA to form depurinating adducts. The resultant apurinic sites in critical genes can generate mutations that may initiate various human cancers. The noncarcinogenic 2-hydroxy CE are oxidized to CE-2,3-Q and form only stable DNA adducts. As reported here, the CE-3,4-Q were bound to DNA in vitro to form the depurinating adduct 4-OHE1(E2)-1(α,β)-N7Gua at 59–213 μmol/mol DNA–phosphate whereas the level of stable adducts was 0.1 μmol/mol DNA–phosphate. In female Sprague–Dawley rats treated by intramammillary injection of E2-3,4-Q (200 nmol) at four mammary glands, the mammary tissue contained 2.3 μmol 4-OHE2-1(α,β)-N7Gua/molDNA–phosphate. When 4-OHE1(E2) were activated by horseradish peroxidase, lactoperoxidase, or cytochrome P450, 87–440 μmol of 4-OHE1(E2)-1(α, β)-N7Gua was formed. After treatment with 4-OHE2, rat mammary tissue contained 1.4 μmol of adduct/mol DNA–phosphate. In each case, the level of stable adducts was negligible. These results, complemented by other data, strongly support the hypothesis that CE-3,4-Q are endogenous tumor initiators.
Cancer is a disease that begins with mutation of critical regulatory genes: oncogenes and tumor suppressor genes (1). The fundamental question is, “How do these cancer-causing mutations arise?” The etiology of most human cancers remains unknown, but we think that the major carcinogenic risk to humans is represented by endogenous carcinogens.
Historically, research in chemical and hormonal carcinogenesis has proceeded on two separate tracks. Chemical carcinogenesis became an important branch of cancer research in the late 1960s when James and Elizabeth Miller postulated the basic principles that constitute the foundation of this discipline (2, 3). They hypothesized that covalent binding of chemicals to cellular macromolecules, DNA, RNA and proteins, is the first critical step in the multi-stage process leading to tumor formation. The common feature of the species reacting with cellular macromolecules is their electrophilicity. This basic discovery directed researchers to focus their studies on DNA adducts produced by various carcinogens. Identification and quantitation of these adducts has led to an understanding of why and how they can initiate cancer (4, 5).
The results of our comprehensive carcinogenesis studies on polycyclic aromatic hydrocarbons (5, 6) have enabled us to conclude that depurinating hydrocarbon–DNA adducts, i.e., adducts lost from DNA by cleavage of the bond between the deoxyribose and the DNA base, are responsible for the critical mutations found in the Harvey-ras oncogene of hydrocarbon-induced mouse skin papillomas (4). These mutations are generated by mis-replication of unrepaired apurinic sites derived from loss of depurinating adducts. This relationship led us to postulate that oxidation of catechol estrogens (CE) to CE quinones (CE-Q) results in electrophilic intermediates that also covalently bind to DNA and form depurinating adducts. The resultant apurinic sites in critical genes can generate mutations that initiate cancer. Thus, the genotoxic effects of specific electrophilic metabolites of estrogens may be at the origin of many human cancers, including breast, endometrium, ovary, prostate, and, possibly, brain cancers.
Hormonal carcinogenesis, in particular carcinogenesis by estrogens, mainly has been investigated by studying the effects of these compounds on increased cell proliferation (7–9). In such research, mitogenic effects generated by receptor-mediated processes are considered critical in tumor promotion and progression. Although hormonal effects can mediate cell proliferation, a genotoxic event is necessary to produce mutations, the permanent genetic changes at the origin of cancer (1). Evidence that estrogens can, indeed, exert their roles as tumor initiators is provided by their well documented carcinogenicity in animal models, in particular the Syrian golden hamster, in which kidney tumors are induced without detection of “spontaneous” tumors in untreated animals (10–13).
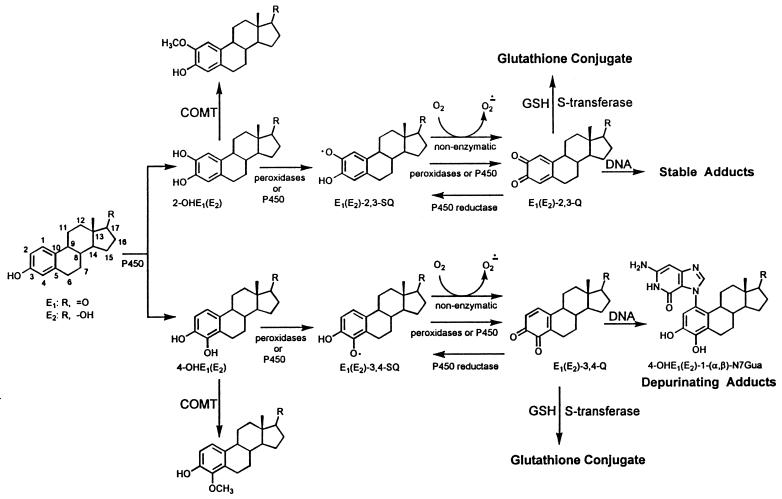
The estrogens 17β-estradiol (E2) and estrone (E1) are metabolized via two major pathways: 16α-hydroxylation (not shown) and formation of CE, the 2-hydroxy and 4-hydroxy derivatives (Fig. 1) (14, 15). Generally, these two CE are mainly inactivated by O-methylation catalyzed by catechol-O-methyltransferases. Although other possible conjugations of CE, such as glucuronidation and sulfation (not shown) may also play a role, if production of these conjugates is incomplete, CE may be oxidized to semiquinones (CE-SQ) and CE-Q (Fig. 1). CE-Q may conjugate with glutathione, catalyzed by S-transferase, or be reduced to CE by quinone reductase (not shown). If these two inactivating processes are incomplete, CE-2,3-Q react with DNA to form stable adducts that remain in DNA unless repaired (16, 17). As presented in this article, CE-3,4-Q, if not inactivated, react with DNA to form depurinating adducts, more specifically two rotational conformers bound to the N-7 of guanine (Gua) (17); these adducts are lost from DNA by cleavage of the glycosidic bond, leaving apurinic sites.
Figure 1.
Metabolism and DNA adducts of estrogens. COMT, catechol-O-methyltransferase.
We hypothesize that formation of N7Gua adducts by reaction of endogenous CE-3,4-Q with DNA is a tumor-initiating event in a number of human cancers. In fact, when tested in male Syrian golden hamsters, 4-hydroxy catechol estrogens (4-OHE) are carcinogenic to the kidney whereas 2-hydroxy catechol estrogens (2-OHE) are not (Table 1) (18, 19). As a test of this hypothesis, we have studied the formation of N7Gua depurinating adducts from reaction of CE-3,4-Q or activated 4-OHE with DNA both in vitro and in vivo.
Table 1.
Carcinogenicity of catechol estrogens in the kidney of male Syrian golden hamsters
| Compound | Tumors, n/animals, n
|
|
|---|---|---|
| Liehr et al. (18) | Li and Li (19) | |
| 4-OHE1 | 2/6 (33%) | |
| 4-OHE2 | 4/5 (80%) | 5/5 (100%) |
| 2-OHE1 | 0/6 | |
| 2-OHE2 | 0/5 | 0/6 |
MATERIALS AND METHODS
Chemicals.
Synthesis of 4-OHE1, 4-OHE2, 2-OHE1, 2-OHE2, and the corresponding quinones was conducted according to Dwivedy et al. (16). E1, E2, and biochemicals were purchased from Sigma; DNA was purchased from Pharmacia; and benzo[a]pyrene was purchased from Eastman.
Instrumentation.
HPLC. Preparative HPLC was conducted on a Waters 600E solvent delivery system equipped with a 484 tunable absorbance detector operating at 290 nm. Analytical HPLC using fluorescence detection was conducted on a Waters 600E solvent delivery system equipped with a 700 WISP autoinjector, a 996 photodiode array detector, and a 474 scanning fluorescence detector, all interfaced to a NEC Powermate 486/33i computer. Analytical HPLC using electrochemical detection was conducted on a Scientific Systems (State College, PA) Model 300 LC pump equipped with an ESA (Chelmsford, MA) Coulochem II electrochemical detector set at 600 mV and monitored on a Hewlett–Packard HP3396A integrator.
Mass Spectrometry (MS).
(i) Electrospray ionization-MS mass spectra were collected by using a prototype VG ZAB-T four-sector mass spectrometer of BEBE configuration at the National Institutes of Health Mass Spectrometry Research Center, Washington University, as described (20). For generating a calibration curve to quantify the in vitro adduct, 30 scans over 60 Da at a rate of 35 s/decay were signal averaged, and the background from the same number of scans was subtracted. The solvents methanol/water/acetic acid (50:49:1) and acetonitrile/water (1:1) for positive and negative mode, respectively, were infused by a syringe pump at a rate of 10 μL/min. A 5-μl aliquot of a 4-OHE1-1(α, β)-N7Gua reference sample with concentrations ranging from 25 fmol/μl to 10 pmol/μl was flow injected via a six-port Rheodyne (Cotati, CA) 7125 loop injector.
(ii) Matrix-Assisted Laser Desorption Ionization-Postsource Decay (MALDI-PSD). Approximately 50 MALDI-PSD (21) spectra were averaged with a PerSeptive BioSystems (Framingham, MA) Voyager single-stage reflectron time-of-flight mass spectrometer at the National Institutes of Health Mass Spectrometry Research Center, Washington University, as described. The sample was dissolved in methanol, and a 1-μl aliquot was added to 1 μl of the matrix solution (α-cyano-4-hydroxycinnamic acid) dissolved in acetonitrile/water/trifluoroacetic acid (50:49:1). A 1-μl aliquot of the resulting solution was deposited on the sample plate.
(iii) Fast Atom Bombardment MS/MS. Tandem mass spectra of the depurinating adduct formed in vivo were obtained at the Nebraska Center for Mass Spectrometry by using a Micromass (Manchester, England) AutoSpec high resolution magnetic sector mass spectrometer. The instrument was equipped with an orthogonal acceleration time-of-flight serving as the second mass spectrometer in the tandem experiment. Xenon was admitted to the collision cell at a level to attenuate the precursor ion signal by 75%. Data acquisition and processing were accomplished using opus software that was provided by the manufacturer (Microcasm). Samples were dissolved in 5–10 μl of methanol; 1-μl aliquots were placed on the sample probe tip along with 1 μl of a 1:1 mixture of glycerol/thioglycerol. Protonated molecules of m/z 966 were produced using fast atom bombardment with a Cs+ ion gun operated at 25 kV.
Carcinogenicity of Catechol Estrogens and Their Quinones in B6C3F1 Mice.
Litters of B6C3F1 mice were obtained from cross-breeding NCI-C3H male mice and NCI-C57-Black female mice (NCI, Frederick, MD). Pups were divided into 13 groups of 20–35 of each sex. Groups were treated by i.p. injection on 4 consecutive days, starting at 12 days of age, with the compound in 10 μl of trioctanoin/dimethyl sulfoxide (DMSO) (95:5). A positive control group was treated with benzo[a]pyrene. One negative control group was injected with the solvent, and another was left untreated. Mice were maintained for approximately 18 months and then killed. Complete necropsies were performed, and tissues were fixed in 10% buffered formalin. Liver and grossly abnormal tissues, including grossly identified tumors, were formalin-fixed, sectioned, stained with hematoxylin and eosin, and examined histopathologically. Statistical analysis of the experiment was conducted using Fisher’s exact test (22) for the number of tumor-bearing animals.
Covalent Binding of 4-Hydroxy Catechol Estrogens and Their Quinones to DNA.
E1-3,4-Q or E2-3,4-Q (1 mg/50 μl of DMSO) was mixed with 5 ml of 6 mM calf thymus DNA in 0.067 M sodium-potassium phosphate (pH 7.0) and incubated for 2 h at 37°C. DNA was precipitated with two volumes of ethanol, and the supernatant was analyzed for depurinating adducts. The DNA was redissolved in 15 mM NaCl/1.5 mM sodium citrate, the concentration was determined by absorbance at 260 nm, and DNA was used for analysis of stable adducts by 32P-postlabeling (16). 4-OHE1 was bound to DNA in reactions catalyzed by horseradish peroxidase (HRP) or lactoperoxidase (LP) in the presence of H2O2 or with phenobarbital-induced rat liver microsomal cytochrome P450 (23) in the presence of cumene hydroperoxide. In each 15-ml peroxidase-catalyzed reaction, mixtures containing 3 mM calf thymus DNA in 0.067 M sodium–potassium phosphate (pH 7.0), 4-OHE1 (1 mg/50 μl DMSO), 0.5 mM H2O2, and 100 μg of HRP (31 units, type VI) or 100 μg of LP (9 units) were incubated for 2 h at 37°C. The H2O2 solution was added at 30-min intervals. For microsome-catalyzed reactions, 15-ml mixtures containing 3 mM calf thymus DNA in 150 mM Tris⋅HCl (pH 7.5), 150 mM KCl, 5 mM MgCl2, 4-OHE1 (1 mg/50 μl DMSO), 1 mg of microsomal protein, and 1 mM cumene hydroperoxide were incubated for 2 h at 37°C. A 1-ml aliquot was used for analysis of stable adducts after purification of the DNA (24). DNA was precipitated from the remaining incubation mixture with 2 vols of ethanol, and the supernatant was used for analysis of depurinating adducts. Control reactions were carried out under identical conditions either with no enzyme or no cofactor.
Binding of 4-Hydroxyestradiol and Estradiol-3,4-Quinone to DNA in Rat Mammary Gland.
Groups of six 7-week-old, female Sprague–Dawley rats (Harlan Laboratories, Haslett, IN) were lightly anesthetized with ether, and the mammary region was shaved. They were then treated with 4-OHE2 or E2-3,4-Q by intramammillary injection (with a 27-gauge needle) under the nipple region of the fourth and fifth mammary glands on the right and left sides at a dose per gland of 200 nmol in 20 μl of DMSO. After 8 h of exposure to 4-OHE2 or 2 h of exposure to E2-3,4-Q, the animals were killed, and mammary gland areas were excised. Mammary tissue was minced, pooled, ground in liquid nitrogen, and split into two samples weighing ≈1 and 5 g each. The smaller sample was used for isolation of DNA (24) and for analysis of stable adducts by 32P-postlabeling (16) whereas the larger sample was analyzed for depurinating adducts.
Analysis of Depurinating Adducts by HPLC.
Supernatants from in vitro binding reactions were evaporated to dryness under vacuum, and residues were dissolved in 1.5 ml of dimethylformamide (DMF) and analyzed by HPLC. The mammary gland tissue was subjected to Soxhlet extraction with two different solvents. First, tissue was extracted with refluxing hexane for 24 h to remove fat and other lipophilic compounds, followed by refluxing methanol containing 5% acetic acid to extract depurinating adducts and metabolites. Methanol/acetic acid solvent was removed under vacuum, and the residue was dissolved in 1.5 ml of DMF.
The extracted materials in DMF were initially purified by preparative HPLC on a YMC (Morris Plains, NJ) ODS-AQ 5-μm, 120-A column (20 × 250 mm) at a flow rate of 8 ml/min. After elution for 15 min with 40% methanol in water, a 15-min curvilinear gradient (CV6) to 55% methanol in water was conducted. Elution with 55% methanol in water was held for 15 min; then, a 10-min curvilinear gradient (CV6) to 100% methanol was used and held at 100% methanol for 15 min for a total run time of 60 min. An initial run using standard adduct was conducted to establish the retention time of the 4-OHE1(E2)-1(α, β)-N7Gua depurinating adducts, which was ≈40 min. The crude 1.5-ml DMF extract was then injected and collected at the appropriate retention time, as established by the standard adduct. Solvent from the collected fraction was removed under vacuum, and the residue was dissolved in 0.5 ml of methanol.
For in vitro binding experiments, the depurinating adducts were measured directly. A 20-μl aliquot of the sample in methanol was analyzed using an ESA electrochemical detector with a Supelco ABZ-ODS 5-μm, 120-A column (4.6 × 250 mm) eluted with an isocratic solvent system of 30% acetonitrile in 36 mM NH4H2PO4 adjusted to pH 3.0 at a flow rate of 1 ml/min. Synthesized authentic adducts were used as reference markers. The remaining sample in methanol was dried under vacuum, and the residue was sent for structural analysis by MS.
For in vivo experiments, the HPLC preparative fractions were first derivatized with a fluorescent probe and then analyzed by HPLC with fluorescence detection. A 150-μl DMF solution containing 1 mg of 1-pyrenesulfonyl chloride (Molecular Probes) was added to the dried HPLC preparative fraction, followed by 50 μl of 0.2 M NaHCO3. The sample was heated to 50°C for 30 min and passed through a 0.45-μm HPLC filter. A 20-μl aliquot was then analyzed with fluorescence detection (λ ex, 350 nm; λem, 485 nm) using a YMC ODS-AQ 5-μm, 120-A column (4.6 × 250 mm) at a flow rate of 1 ml/min with a methanol/water gradient. After elution for 5 min with 50% methanol in water, a 50-min curvilinear gradient (CV6) to 100% methanol was conducted and held for 20 min, for a total run time of 70 min. The remaining solution was then dried under vacuum, and the residue was analyzed by MS.
Analysis of Stable Adducts by 32P-Postlabeling.
32P-postlabeling analysis of stable DNA adducts was carried out with 8 μg of DNA on 10 × 13-cm polyethyleneimine–cellulose plates as described earlier (16). Adduct spots were detected by autoradiography, and quantities were determined by liquid scintillation counting of each excised spot.
RESULTS
Carcinogenicity.
The carcinogenicity of 4-OHE and the lack of carcinogenicity of 2-OHE (18, 19) (Table 1) led us to investigate another animal model system in which both CE and CE-Q could be tested. Injection (i.p.) of the estrogens E1 and E2, their 2-OHE and 4-OHE, and their CE-Q into young male and female B6C3F1 mice was selected to examine development of liver tumors (Table 2). Among the CE and their quinones, only E1-3,4-Q was significantly carcinogenic in the liver of male mice; a very low dose (3.7 nmol/g body weight/injection; Table 2) was used because of its high toxicity. Even at this low dose, most mice died shortly after treatments. Of the 10 remaining, six developed 13 tumors, demonstrating that E1-3,4-Q was more tumorigenic than treatment with solvent or no treatment. In addition, E1-3,4-Q was not significantly less tumorigenic than the positive control, benzo[a]pyrene. We do not know why E1-3,4-Q was carcinogenic and highly toxic and E2-3,4-Q was not. None of the other estrogens tested was significantly tumorigenic.
Table 2.
Carcinogenicity of estrogens, catechol estrogens, and quinones in B6C3F1 male mice
| Compound | Dose nmol/g body weight | Effective no. of animals | Total no. of tumors/TBA | TBA, % | Liver tumors
|
No. of mice with other neoplasms | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of tumors/TBA | TBA, % | Carcinomas/TBA | TBA, % | Adenomas/TBA | TBA, % | ||||||
| E2 | 30 | 20 | 6/6 | 30 | 5/5 | 25 | 3/3 | 15 | 2/2 | 10 | 1A |
| 2-OHE2 | 30 | 28 | 13/10 | 36 | 10/9 | 32 | 5/4 | 14 | 5/5 | 18 | 1B, 1C, 1D |
| 4-OHE2 | 30 | 24 | 15/10 | 42 | 13/8 | 33 | 6/5 | 21 | 7/3 | 12 | 2A |
| E22,3-Q | 7.5 | 26 | 10/8 | 31 | 108 | 31 | 6/5 | 19 | 4/3 | 12 | |
| E2-3,4-Q | 7.5 | 21 | 5/4 | 19 | 5/4 | 19 | 4/3 | 14 | 1/1 | 5 | |
| E1 | 30 | 32 | 4/3 | 9 | 3/2 | 6* | 0 | 0† | 3/2 | 6 | 1A |
| 2-OHE1 | 30 | 30 | 11/9 | 30 | 9/7 | 23 | 4/4 | 13 | 5/4 | 13 | 1B, 1E |
| 4-OHE1 | 30 | 33 | 8/8 | 24 | 6/6 | 18 | 5/5 | 15 | 2/2 | 6 | 1A |
| E12,3-Q | 7.5 | 25 | 17/12 | 48 | 14/11 | 44 | 12/9 | 36 | 2/2 | 8 | 2A |
| E1-3,4-Q | 3.7 | 10 | 13/6 | 60 | 13/6 | 60‡ | 13/6 | 60§¶ | 0 | ||
| BP | 60 | 12 | 74/12 | 100 | 68/12 | 100 | 55/11 | 92 | 13/7 | 58 | 4B, 1F |
| Solvent | 19 | 11/7 | 37 | 9/5 | 26 | 5/4 | 21 | 4/2 | 10 | 2A | |
| Untreated | 33 | 11/11 | 33 | 8/8 | 24 | 6/6 | 18 | 2/2 | 6 | 3A | |
Effective number of animals, number of mice that survived after weaning; TBA, tumor-bearing animals; A, lung adenoma; B, lung carcinoma; C, eye papillary carcinoma (sebaceous); D, pancreatic lymph node lymphoma; E, spleen lymphoma; F, malignant lymphoma; BP, benzo[a]pyrene.
The mice treated with E1 had significantly fewer tumors than the untreated mice (P = 0.046).
E1 was significantly less carcinogenic than the solvent (P < 0.016) and no treatment (P < 0.013).
E1-3,4-Q was significantly more tumorigenic than no treatment (P < 0.044).
E1-3,4-Q was significantly more carcinogenic than the solvent (P < 0.047).
E1-3,4-Q vs. BP does not approach the nominal 0.05 P value of the level of significance.
The estrogen metabolites induced very few tumors in female B6C3F1 mice (data not shown). This resistance to estrogen-induced carcinogenesis resembles the previously observed resistance to estrogen-induced kidney tumors in female hamsters (12). In summary, B6C3F1 mice are not a good model for investigating estrogen-induced tumors although the results do indicate the role of 4-OHE in forming ultimate carcinogenic metabolites, namely, CE-3,4-Q.
Covalent Binding of 4-Hydroxy Catechol Estrogens and Their Quinones to DNA in Vitro.
Formation of the depurinating adduct 4-OHE1(E2)-1(α, β)-N7Gua upon reaction of E1(E2)-3,4-Q with dG (17) led us to determine whether or not this adduct is formed by reaction of E1(E2)-3,4-Q with DNA or by activation of 4-OHE with HRP, LP or cytochrome P450. Reaction of E2-3,4-Q with DNA yielded 213 μmol/mol DNA–phosphate of 4-OHE2-1(α,β)-N7Gua whereas E1-3,4-Q produced four times less (Table 3). Both CE-Q afforded significantly more depurinating adducts compared with stable adducts. When 4-OHE2 and 4-OHE1 were activated by HRP in the presence of DNA, 4-OHE2 produced four times more depurinating adduct than 4-OHE1, and the levels of binding were similar to those obtained with the two quinones. Activation of 4-OHE1 by the mammalian LP gave nine times more N7Gua adduct than when HRP was used. Phenobarbital-induced rat liver microsomes activated 4-OHE1 only with cumene hydroperoxide as the cofactor and not NADPH. The microsomal activation resulted in 87 μmol/mol DNA–phosphate of 4-OHE1-1(α,β)-N7Gua.
Table 3.
Reaction of CE-Q and HRP-, LP-, or P450-activated CE with DNA
| Compound | 4-OHE1,2-1(α,β)-N7Gua, μmol/mol DNA-P | Stable adducts, μmol/mol, DNA-P | Depurinating/stable adducts |
|---|---|---|---|
| E2-3,4,-Q | 213 | 0.07 | 3043 |
| HRP-activated 4-OHE2 | 194 | 0.10 | 1940 |
| E1-3,4-Q | 59 | 0.11 | 536 |
| HRP-activated 4-OHE1 | 50 | 0.07 | 714 |
| LP-activated 4-OHE1 | 440 | 0.06 | 7333 |
| PB-microsome/CuOOH-activated 4-OHE1 | 87 | 0.01 | 8700 |
CuOOH, cumene hydroperoxide; HRP, horseradish peroxidase; LP, lactoperoxidase; PB, phenobarbital.
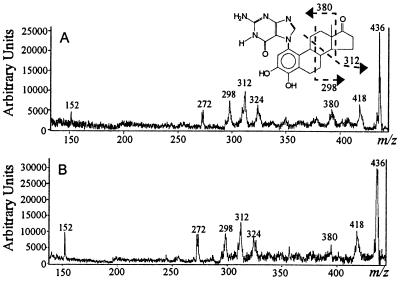
The retention time of the depurinating adduct was not sufficient to prove its identity, and MS was used. A calibration plot (not shown) is linear (correlation coefficient is >0.995) for 4-OHE1-1(α, β)-N7Gua. Adducts from in vitro experiments were detected in both positive and negative mode electrospray ionization-MS. Using the calibration plot, we established that ≈5 pmol was injected into the electrospray ionization source, indicating that ≈30 pmol was isolated from the in vitro experiment. Because negative-ion mode ES is less susceptible to interference, the spectrum obtained in this mode is cleaner than that from the positive-ion mode. The structure of the in vitro adduct that was isolated from the same experiments was confirmed by MALDI-PSD because there was insufficient material for tandem, four-section MS. All characteristic fragment ions in the reference spectrum of 4-OHE1-1(α, β)-N7Gua (Fig. 2B) were in the spectrum obtained for the in vitro adduct (Fig. 2A). Upon MALDI, the [M + H]+ ions decompose to give a PSD spectrum of product ions of m/z 418 (formed by loss of water), m/z 324, 312, 298, and 272 [formed by charge-remote fragmentations that established adduction at the A ring of the steroid (17)], and m/z 152 (protonated Gua, proving that the adduct is a modified Gua).
Figure 2.
MALDI-PSD spectra of the [M + H]+ ions from (A) 4-OHE1-1(α, β)-N7Gua formed in vitro and (B) synthetic reference 4-OHE1-1(α, β)-N7Gua.
Covalent Binding of 4-Hydroxyestradiol and its 3,4-Quinone to DNA in Vivo.
The 4-OHE2-1(α, β)-N7Gua depurinating adduct also was detected in vivo after treatment of female Sprague–Dawley rats with 200 nmol of E2-3,4-Q or 4-OHE2 at each of four mammary glands. The adduct was detected at a level of 2.3 or 1.4 μmol/mol DNA–phosphate for treatment with E2-3,4-Q or 4-OHE2, respectively (Table 4), whereas the level of stable adducts was <0.01 μmol of adduct/mol DNA–phosphate, the limit of detection.
Table 4.
Formation of 4-OHE2-1(α,β)-N7Gua in the mammary gland of Sprague–Dawley rats
| Compound | 4-OHE2-1(α,β)-N7Gua, μmol/mol DNA-P |
|---|---|
| E2-3,4-Q | 2.3 |
| 4-OHE2 | 1.4 |
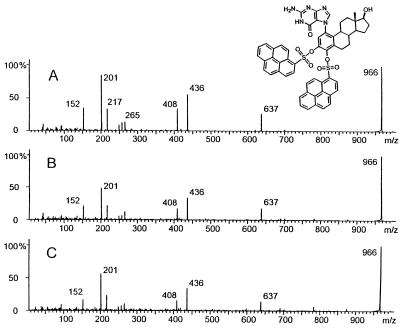
The structure of the depurinating adduct formed in the mammary gland after treatment with E2-3,4-Q or 4-OHE2 was proven by tandem MS of the disulfonylpyrene-derivatized 4-OHE2-1(α, β)-N7Gua adduct, which has an [M + H]+ ion of m/z 966 (Fig. 3A). Three of the abundant fragment ions are distinctly characteristic of portions of the molecule. The product ion of m/z 152 is protonated Gua, that of m/z 201 is the pyrene moiety (C16H9), and that of m/z 265 is SO2-pyrene. The other major ions result from fragmentation processes occurring after a molecular rearrangement. The ion of m/z 637 arises from elimination of the elements of C16H9SO2 that correspond to one of the SO2-pyrene derivatizing groups and an additional unit of SO2. This can only be accomplished if, after elimination of an SO2-pyrene group, the oxygen remaining on the estrogen attacks the C-1 position of the second pyrene, transferring the pyrene to the adjacent oxygen. After rearrangement, the protonated molecule eliminates SO2 to produce the ion of m/z 637, which then eliminates pyrene to produce the ion of m/z 436. Elimination of CO from the ion of m/z 436 results in the ion of m/z 408. Further evidence of this rearrangement process is the ion of m/z 217, which is [pyrene + O].
Figure 3.
MS/MS spectra of m/z 966 ions produced by fast atom bombardment of (A) 43 pmol of disulfonylpyrene-derivatized 4-OHE2-1(α, β)-N7Gua, (B) derivatized extract from mammary tissue of rats treated with E2-3,4-Q, and (C) derivatized extract from mammary tissue of rats treated with 4-OHE2. The abundance of fragment ions from 0–650 is amplified 3-fold with respect to the parent ion (m/z 966).
The product–ion spectra of [M + H]+ ions of m/z 966 obtained from the extracts of mammary tissues of rats treated with E2-3,4-Q and 4-OHE2 in separate experiments are shown in Fig. 3 B and C, respectively. Both spectra agree with that of the standard; all characteristic fragment ions are observed in both spectra. From the signal-to-noise ratio of the spectra, we estimate that the isolates contained between 5 and 30 pmol of adduct.
The results of these in vitro and in vivo studies demonstrate that CE-3,4-Q covalently bind to DNA to form 4-OHE1(E2)-1(α, β)-N7Gua. Peroxidases and cytochrome P450 can activate 4-OHE to form the same depurinating adduct, which is also found when rat mammary glands are treated with 4-OHE2 or its corresponding quinone. In each case, the amount of depurinating adducts is several orders of magnitude greater than that of stable adducts.
DISCUSSION
The depurinating adduct 4-OHE1(E2)-1(α, β)-N7Gua was formed in vitro and in vivo by reaction of E1(E2)-3,4-Q with DNA and after activation of 4-OHE by cytochrome P450 and peroxidases (Tables 3 and 4). CE-Q derived from 4-OHE and 2-OHE react differently with nucleosides and DNA because of their distinctive chemical properties (17). CE-2,3-Q bind to the exocyclic amino groups of dA and dG to form adducts that retain the deoxyribose moiety whereas CE-3,4-Q bind exclusively to the N-7 of Gua, resulting in destabilization of the glycosidic bond and subsequent depurination. CE-2,3-Q form 10–50 times higher levels of stable DNA adducts than CE-3,4-Q (16). CE-3,4-Q, however, give two to three orders of magnitude higher levels of depurinating than stable adducts (Table 3). Thus, CE-2,3-Q produce stable DNA adducts (16), and the corresponding CE do not induce kidney tumors in Syrian golden hamsters (18, 19) whereas CE-3,4-Q form predominantly 4-OHE1(E2)-1(α, β)-N7Gua, and E1-3,4-Q is carcinogenic in mouse liver (Table 2); in addition, 4-OHE are carcinogenic in Syrian golden hamsters (18, 19). These data strongly support the view that CE-3,4-Q are endogenous ultimate carcinogens.
To corroborate this evidence, synthetic nonsteroidal estrogens, which are carcinogenic in animals (10, 11), were studied. Among these, diethylstilbestrol is also a human carcinogen (10, 11, 25). The first nonsteroidal estrogen investigated was hexestrol, the derivative of diethylstilbestrol that is hydrogenated at the C-C double bond. This compound is carcinogenic in Syrian golden hamsters (13, 26). The major metabolite of hexestrol is its catechol (26, 27), which can be metabolically converted to its catechol quinone. Hexestrol quinone has the same chemical and biochemical properties as CE-3,4-Q; namely, it specifically forms an N7Gua adduct after reaction with dG or DNA. Furthermore, the hexestrol catechol is activated by HRP, LP, or cytochrome P450 to form an N7Gua adduct (ref. 28 and S. T. Jan, P.D.D., D.E.S., R.R., M.L.G., E.G.R., and E.L.C., unpublished work).
There is additional evidence that 4-OHE are critical intermediates in the pathway leading to estrogen-induced cancer, as reviewed by Yager and Liehr (29). Estrogen 4-hydroxylase activity has been identified in hamster kidney (30, 31) and other organs prone to estrogen-induced cancers, such as rat pituitary (32) and mouse uterus (33). In humans, predominant 4-hydroxylase activity has been detected in microsomes of uterine myometrium and benign uterine leiomyomas (34) and in benign and malignant breast tumors (35, 36). One enzyme with specific 4-hydroxylase activity has been identified as cytochrome P450 1B1, which is found in 15 human tissues, including mammary, ovary, and adrenal (37, 38). The 4-hydroxylase activity found in MCF-7 human breast cancer cells is induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an environmental pollutant, and correlates with increased formation of 4-OHE2 (39). Homologous P450 1B1 has been isolated from the liver of Sprague–Dawley rats induced with TCDD (40). The mRNA levels in kidney and liver from TCDD-induced female rats were greater than those in TCDD-induced male rats. The potent hepatocarcinogenic activity of TCDD in female, but not male, rats suggests involvement of 4-OHE2 in the induction of these tumors.
Redox cycling (41, 42) generated by enzymatic reduction of CE-Q to CE-SQ and subsequent autoxidation back to CE-Q by dioxygen forms superoxide radicals (Fig. 1) and, subsequently, hydroxy radicals. In turn, hydroxy radicals can produce lipid hydroperoxides (43) and DNA damage (42, 44–46). The DNA damage includes C-8 hydroxylation of dA and dG and has been found to be abundant in mammary DNA from breast cancer patients (47) and prostate DNA from prostate cancer patients (48). Thus, redox cycling of CE-Q/CE-SQ/CE-Q could contribute to the initiating process by (i) enhancing oxidation of CE to CE-Q, thus rendering less competitive the process of protection by conjugating enzymes (Fig. 1) and (ii) forming excessive 8-hydroxy dA and dG that can increase critical mutagenic events.
Induction of 4-hydroxylase enzymes that would produce increased levels of 4-OHE, accompanied by insufficient conjugation of the proximate and ultimate metabolites, can lead to formation of CE-3,4-Q, which react with DNA to form depurinating N7Gua adducts by Michael addition (Fig. 1). Loss of these adducts generates apurinic sites in DNA, which have high potential to produce mutations in critical genes. This series of events can initiate cancer in a variety of human tissues. After tumor initiation by CE-3,4-Q, hormone receptor-mediated processes would play a major role in the promotion and progression phases of tumor development.
Acknowledgments
We thank Dr. P. Mulder for synthesizing the CE. We acknowledge support from U.S. Public Health Service Grant PO1 CA49210 and a supplement to Eppley Institute Core Grant CA36727 from the National Cancer Institute. Core support was provided to Washington University by Grant P41RR00954.
ABBREVIATIONS
- CE
catechol estrogen(s)
- CE-Q
catechol estrogen quinone(s)
- DMF
dimethylformamide
- DMSO
dimethyl sulfoxide
- E1
estrone
- E2
17β-estradiol
- Gua
guanine
- HRP
horseradish peroxidase
- LP
lactoperoxidase
- MALDI-PSD
matrix-assisted laser desorption ionization-postsource decay
- MS
mass spectrometry
- 2-OHE
2-hydroxy estradiol and estrone
- 4-OHE
4-hydroxy estradiol and estrone
References
- 1.Weinberg R A. Sci Am. 1996;275:62–77. doi: 10.1038/scientificamerican0996-62. [DOI] [PubMed] [Google Scholar]
- 2.Miller J A. Cancer Res. 1970;30:559–576. [PubMed] [Google Scholar]
- 3.Miller E C, Miller J A. Cancer. 1981;47:2327–2345. doi: 10.1002/1097-0142(19810515)47:10<2327::aid-cncr2820471003>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti D, Pelling J C, Cavalieri E L, Rogan E G. Proc Natl Acad Sci USA. 1995;92:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalieri E, Rogan E. In: The Handbook of Environmental Chemistry: PAHs and Related Compounds. Neilson A H, editor. Vol. 3. Heidelberg: Springer; 1997. , in press. [Google Scholar]
- 6.Cavalieri E L, Rogan E G. Pharmacol Ther. 1992;55:183–199. doi: 10.1016/0163-7258(92)90015-r. [DOI] [PubMed] [Google Scholar]
- 7.Preston-Martin S, Pike M C, Ross R K, Jones P A, Henderson B E. Cancer Res. 1990;50:7415–7421. [PubMed] [Google Scholar]
- 8.Nandi S, Guzman R C, Yang J. Proc Natl Acad Sci USA. 1995;92:3650–3657. doi: 10.1073/pnas.92.9.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ethier S P. J Natl Cancer Inst. 1995;87:964–973. doi: 10.1093/jnci/87.13.964. [DOI] [PubMed] [Google Scholar]
- 10.International Agency for Research on Cancer: Monographs on the Evaluation of Carcinogenic Risks to Humans (1979) 21, 279–362. [PMC free article] [PubMed]
- 11.International Agency for Research on Cancer: Monographs on the Evaluation of Carcinogenic Risks to Humans (1987) 7, Suppl., 272–310. [PMC free article] [PubMed]
- 12.Kirkman H. Natl Cancer Inst Monogr. 1959;1:1–59. [PubMed] [Google Scholar]
- 13.Li J J, Li S A, Klicka J K, Parsons J A, Lam L K T. Cancer Res. 1983;43:5200–5204. [PubMed] [Google Scholar]
- 14.Ball, P. & Knuppen, R. (1980) Acta Endocrinol. (Copenhagen) 93, Suppl. 232, 1–127. [PubMed]
- 15.Martucci C P, Fishman J. Pharmacol Ther. 1993;57:237–257. doi: 10.1016/0163-7258(93)90057-k. [DOI] [PubMed] [Google Scholar]
- 16.Dwivedy I, Devanesan P, Cremonesi P, Rogan E, Cavalieri E. Chem Res Toxicol. 1992;5:828–833. doi: 10.1021/tx00030a016. [DOI] [PubMed] [Google Scholar]
- 17.Stack D, Byun J, Gross M L, Rogan E G, Cavalieri E. Chem Res Toxicol. 1996;9:851–859. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- 18.Liehr J G, Fang W F, Sirbasku D A, Ari-Ulubelen A. J Steroid Biochem. 1986;24:353–356. doi: 10.1016/0022-4731(86)90080-4. [DOI] [PubMed] [Google Scholar]
- 19.Li J J, Li S A. Fed Proc. 1987;46:1858–1863. [PubMed] [Google Scholar]
- 20.Byun, J., Gooden, J., Ramanathan, R., Li, K., Cavalieri, E. & Gross, M. L. (1997) J. Am. Soc. Mass Spectrom, in press. [DOI] [PubMed]
- 21.Kaufmann R, Kirsch D, Spengler B. Int J Mass Spectrom Ion Process. 1994;131:355–385. [Google Scholar]
- 22.Armitage P. Statistical Methods in Medical Research. Oxford: Blackwell Scientific; 1994. p. 112. [Google Scholar]
- 23.Wong A K L, Cavalieri E L, Rogan E G. Biochem Pharmacol. 1985;35:1583–1588. doi: 10.1016/0006-2952(86)90128-0. [DOI] [PubMed] [Google Scholar]
- 24.Bodell W J, Devanesan P D, Rogan E G, Cavalieri E L. Chem Res Toxicol. 1989;2:312–315. doi: 10.1021/tx00011a008. [DOI] [PubMed] [Google Scholar]
- 25.Herbst A L, Ulfeder H, Poskanzer D C. N Engl J Med. 1971;284:878–881. doi: 10.1056/NEJM197104222841604. [DOI] [PubMed] [Google Scholar]
- 26.Liehr J G, Ballatore A M, Dague B B, Ulubelen A A. Chem Biol Interact. 1985;55:157–176. doi: 10.1016/s0009-2797(85)80125-3. [DOI] [PubMed] [Google Scholar]
- 27.Metzler M, McLachlan J A. Adv Exp Med Biol. 1981;136A:829–837. doi: 10.1007/978-1-4757-0674-1_65. [DOI] [PubMed] [Google Scholar]
- 28.Jan S T, Devanesan P D, Rogan E G, Cavalieri E L. Proc Am Assoc Cancer Res. 1996;37:120. [Google Scholar]
- 29.Yager J D, Liehr J G. Annu Rev Pharmacol Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- 30.Weisz J, Bui Q D, Roy D, Liehr J G. Endocrinology. 1992;131:655–661. doi: 10.1210/endo.131.2.1386303. [DOI] [PubMed] [Google Scholar]
- 31.Zhu B T, Bui Q D, Weisz J, Liehr J G. Endocrinology. 1994;135:1772–1779. doi: 10.1210/endo.135.5.7956900. [DOI] [PubMed] [Google Scholar]
- 32.Bui Q D, Weisz J. Pharmacology. 1988;36:356–364. doi: 10.1159/000138406. [DOI] [PubMed] [Google Scholar]
- 33.Bunyagidj C, McLachlan J A. J Steroid Biochem. 1988;31:295–801. doi: 10.1016/0022-4731(88)90288-9. [DOI] [PubMed] [Google Scholar]
- 34.Liehr J G, Ricci M J, Jefcoate C R, Hannigan E V, Hokanson J A, Zhu B T. Proc Natl Acad Sci USA. 1995;92:9220–9224. doi: 10.1073/pnas.92.20.9220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liehr J G, Ricci M J. Proc Natl Acad Sci USA. 1996;93:3294–3296. doi: 10.1073/pnas.93.8.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castagnetta L A, Granata O M, Arcuri F P, Polito L M, Rosati F, Cartoni G P. Steroids. 1992;57:437–443. doi: 10.1016/0039-128x(92)90097-s. [DOI] [PubMed] [Google Scholar]
- 37.Savas U, Bhattacharya K K, Christou M, Alexander D L, Jefcoate C R. J Biol Chem. 1994;269:14905–14911. [PubMed] [Google Scholar]
- 38.Hayes C L, Spink D C, Spink B C, Cao J Q, Walker N J, Sutter T R. Proc Natl Acad Sci USA. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spink D C, Hayes G L, Young N R, Christou M, Sutter T R, Jefcoate C R, Gierthy J F. J Steroid Biochem Mol Biol. 1994;51:251–258. doi: 10.1016/0960-0760(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 40.Walker N J, Gastel J A, Costa L T, Clark G C, Lucier G W, Sutter T R. Carcinogenesis. 1995;16:1319–1327. doi: 10.1093/carcin/16.6.1319. [DOI] [PubMed] [Google Scholar]
- 41.Liehr J G, Ulubelen A A, Strobel H W. J Biol Chem. 1986;261:16865–16870. [PubMed] [Google Scholar]
- 42.Liehr J G, Roy D. Free Radical Biol Med. 1990;8:415–423. doi: 10.1016/0891-5849(90)90108-u. [DOI] [PubMed] [Google Scholar]
- 43.Wang M Y, Liehr J G. Carcinogenesis. 1995;16:1941–1945. doi: 10.1093/carcin/16.8.1941. [DOI] [PubMed] [Google Scholar]
- 44.Han X, Liehr J G. Carcinogenesis. 1994;54:5515–5517. [PubMed] [Google Scholar]
- 45.Nutter L M, Ngo E O, Abul-Hajj Y J. J Biol Chem. 1991;7:23–28. [PubMed] [Google Scholar]
- 46.Nutter L M, Wu Y-Y, Ngo E O, Sierra E E, Gutierrez P L, Abul-Hajj Y J. Chem Res Toxicol. 1994;7:23–28. doi: 10.1021/tx00037a004. [DOI] [PubMed] [Google Scholar]
- 47.Malins D C, Holmes E H, Polissar N L, Gunselman S J. Cancer. 1993;71:3036–3043. doi: 10.1002/1097-0142(19930515)71:10<3036::aid-cncr2820711025>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 48.Malins D C, Polissar N L, Gunselman S J. Proc Natl Acad Sci USA. 1997;94:259–264. doi: 10.1073/pnas.94.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]