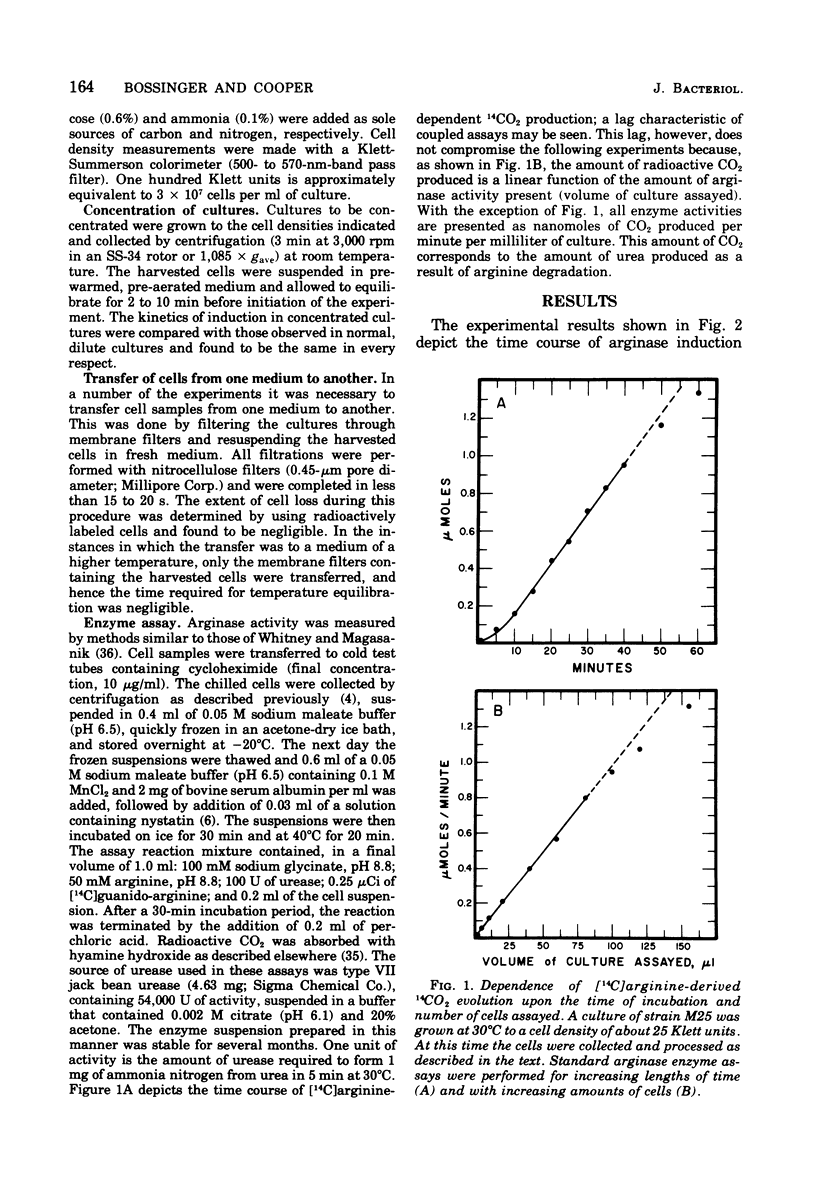
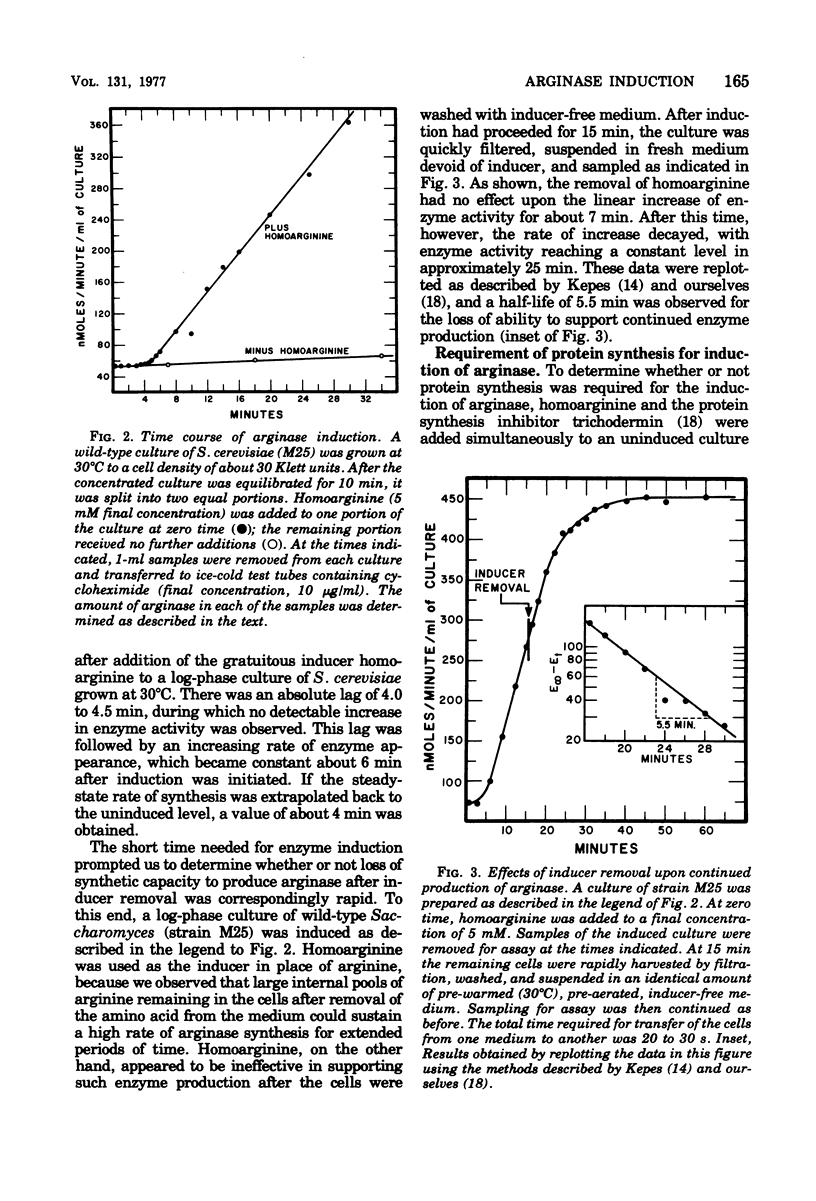
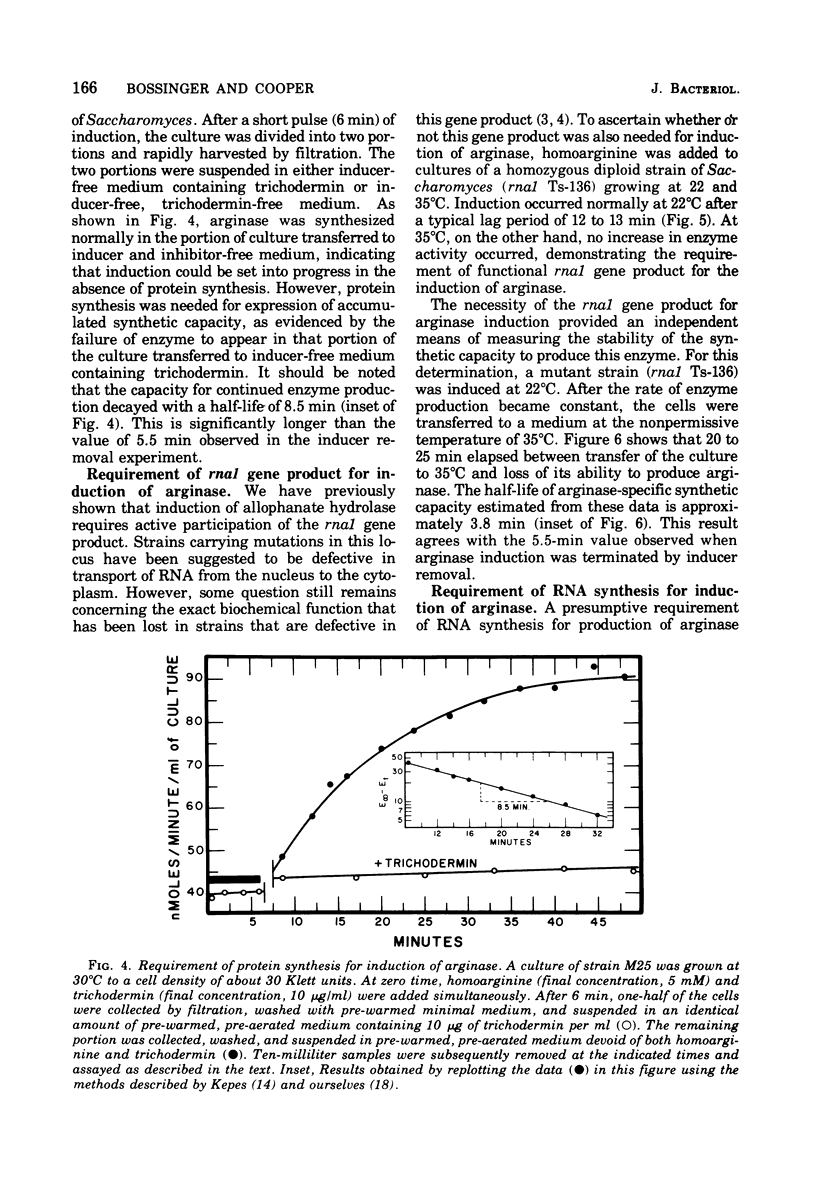
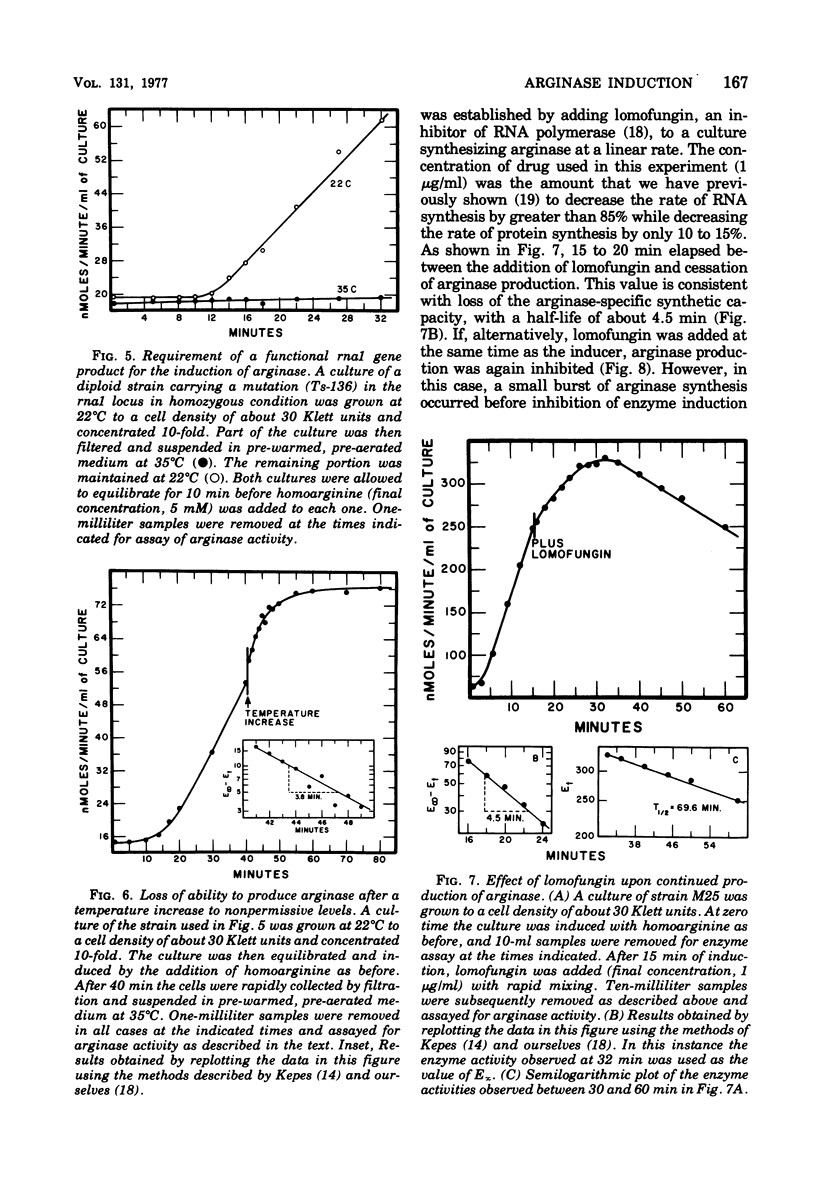
Abstract
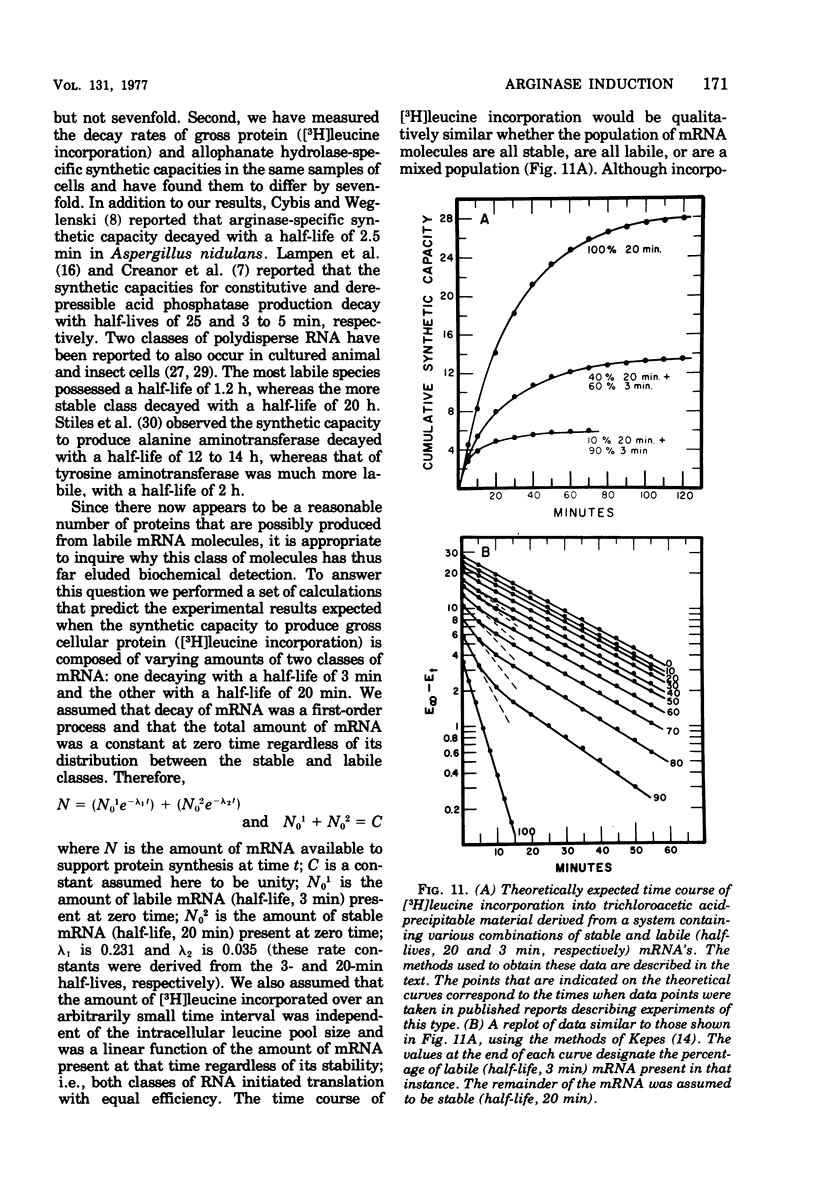
Arginase, the enzyme responsible for arginine degradation in Saccharomyces cerevisiae, is an inducible protein whose inhibition of ornithine carbamoyl-transferase has been studied extensively. Mutant strains defective in the normal regulation of arginase production have also been isolated. However, in spite of these studies, the macromolecular biosynthetic events involved in production of arginase remain obscure. We have, therefore, studied the requirements of arginase induction. We observed that: (i) 4 min elapsed between the addition of inducer (homoarginine) and the appearance of arginase activity at 30 degrees C; (ii) induction required ribonucleic acid synthesis and a functional rna1 gene product; and (iii) production of arginase-specific synthetic capacity occurred in the absence of protein synthesis but could be expressed only when protein synthesis was not inhibited. Termination of induction by inducer removal, addition of the ribonucleic acid synthesis inhibitor lomofungin, or resuspension of a culture of organisms containing temperature-sensitive rna1 gene products in a medium at 35 degrees C resulted in loss of ability for continued arginase synthesis with half-lives of 5.5, 3.8, and 4.5 min, respectively. These and other recently published data suggest that a variety of inducible or repressible proteins responding rapidly to the environment may be derived from labile synthetic capacities, whereas constitutively produced proteins needed continuously throughout the cell cycle may be derived from synthetic capacities that are significantly more stable.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bechet J., Wiame J. M. Indication of a specific regulatory binding protein for ornithinetranscarbamylase in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1965 Nov 8;21(3):226–234. doi: 10.1016/0006-291x(65)90276-7. [DOI] [PubMed] [Google Scholar]
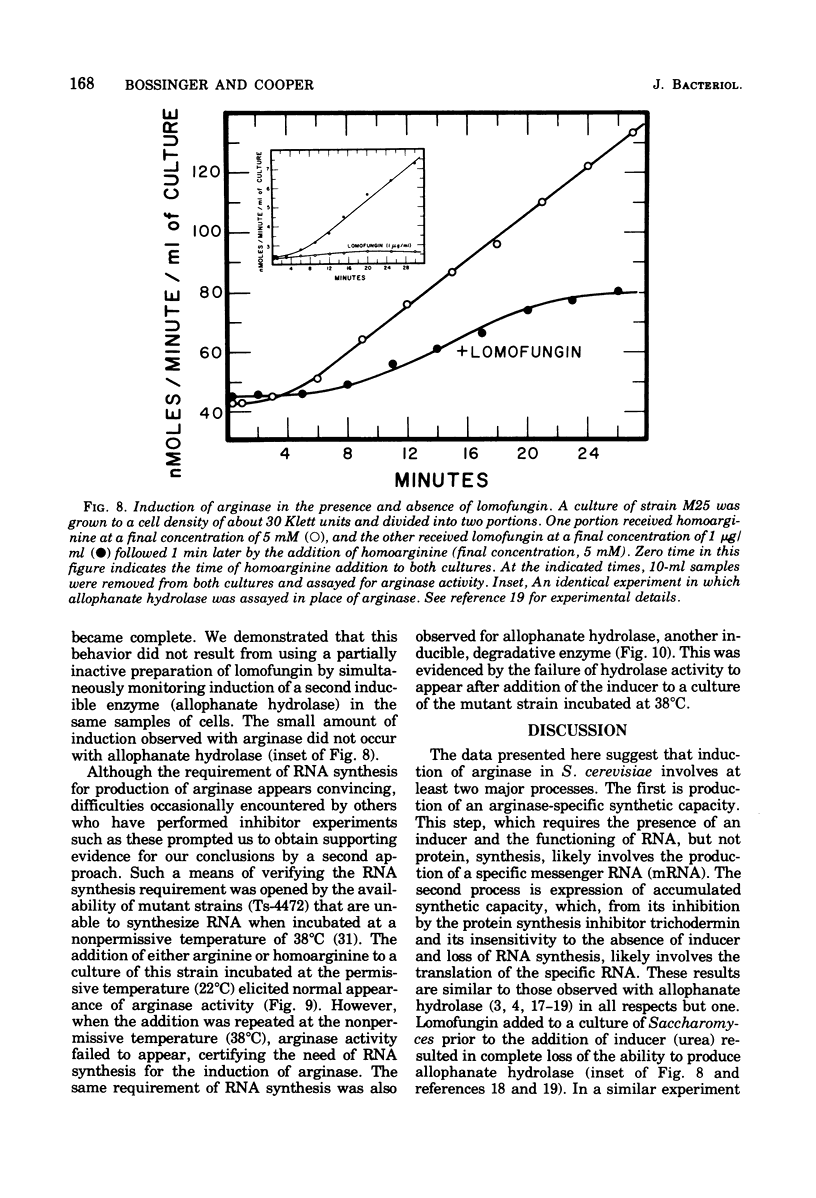
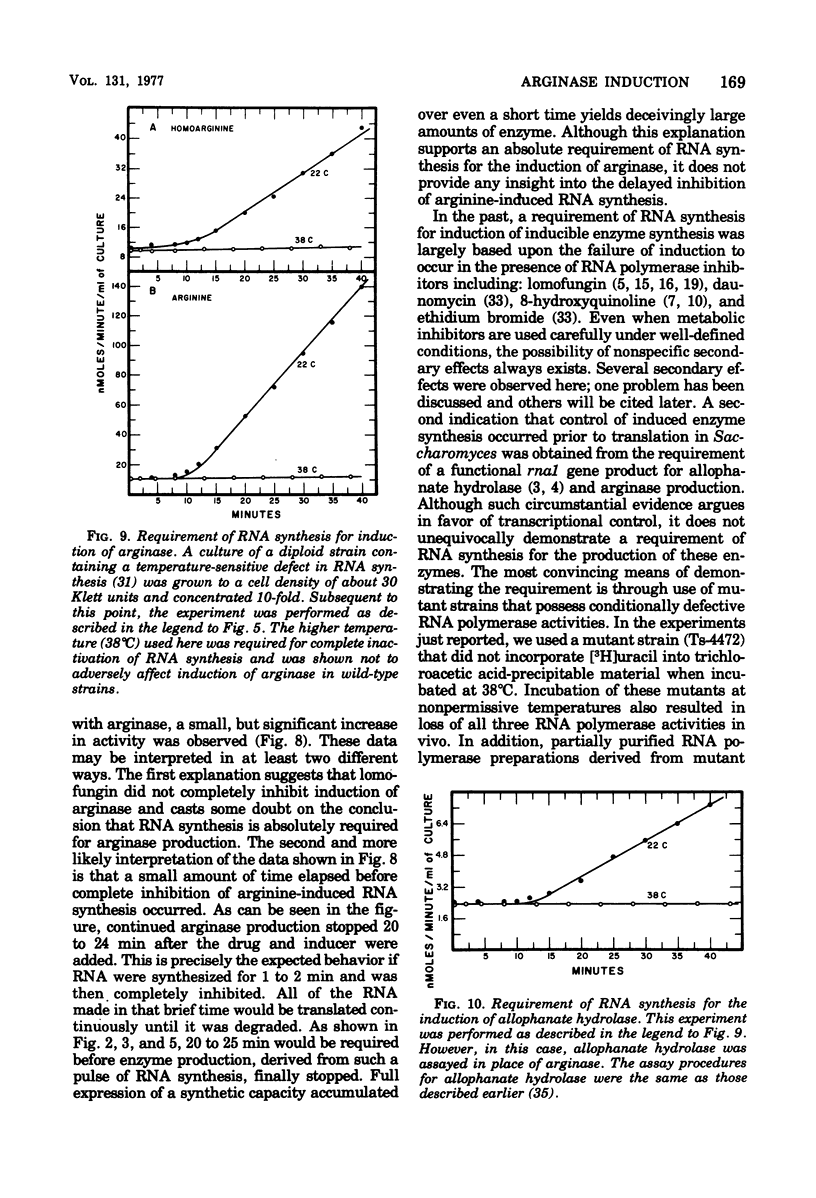
- Bossinger J., Cooper T. G. Execution times of macromolecular synthetic processes involved in the induction of allophanate hydrolase at 15 degrees C. J Bacteriol. 1976 Oct;128(1):498–501. doi: 10.1128/jb.128.1.498-501.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger J., Cooper T. G. Sequence of molecular events involved in induction of allophanate hydrolase. J Bacteriol. 1976 Apr;126(1):198–204. doi: 10.1128/jb.126.1.198-204.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M., Davies J. E., Jimenez A. Inhibition by lomofungin of nucleic acid and protein synthesis in Saccharomyces cerevisiae. FEBS Lett. 1973 Jun 1;32(2):277–280. doi: 10.1016/0014-5793(73)80852-x. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Lawther R. P. Induction of the allantoin degradative enzymes in Saccharomyces cerevisiae by the last intermediate of the pathway. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2340–2344. doi: 10.1073/pnas.70.8.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creanor J., May J. W., Mitchison J. M. The effect of 8-hydroxyquinoline on enzyme synthesis in the fission yeast Schizosaccharomyces pombe. Eur J Biochem. 1975 Dec 15;60(2):487–493. doi: 10.1111/j.1432-1033.1975.tb21027.x. [DOI] [PubMed] [Google Scholar]
- Cybis J., Weglenski P. Arginase induction in Aspergillus nidulans. The appearance and decay of the coding capacity of messenger. Eur J Biochem. 1972 Oct;30(2):262–268. doi: 10.1111/j.1432-1033.1972.tb02094.x. [DOI] [PubMed] [Google Scholar]
- Dubois E., Grenson M., Wiame J. M. Release of the "ammonia effect" on three catabolic enzymes by NADP-specific glutamate dehydrogenaseless mutations in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1973 Feb 20;50(4):967–972. doi: 10.1016/0006-291x(73)91500-3. [DOI] [PubMed] [Google Scholar]
- Fraser R. S. Turnover of polyadenylated messenger RNA in fission yeast. Evidence for the control of protein synthesis at the translational level. Eur J Biochem. 1975 Dec 15;60(2):477–486. doi: 10.1111/j.1432-1033.1975.tb21026.x. [DOI] [PubMed] [Google Scholar]
- Greenberg J. R. High stability of messenger RNA in growing cultured cells. Nature. 1972 Nov 10;240(5376):102–104. doi: 10.1038/240102a0. [DOI] [PubMed] [Google Scholar]
- Hutchison H. T., Hartwell L. H., McLaughlin C. S. Temperature-sensitive yeast mutant defective in ribonucleic acid production. J Bacteriol. 1969 Sep;99(3):807–814. doi: 10.1128/jb.99.3.807-814.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes N. E., Phillips S. L. Turnover of polyadenylate-containing ribonucleic acid in Saccharomyces cerevisiae. J Bacteriol. 1976 Feb;125(2):595–600. doi: 10.1128/jb.125.2.595-600.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEPES A. KINETICS OF INDUCED ENZYME SYNTHESIS. DETERMINATION OF THE MEAN LIFE OF GALACTOSIDASE-SPECIFIC MESSENGER RNA. Biochim Biophys Acta. 1963 Oct 15;76:293–309. [PubMed] [Google Scholar]
- Klo S. C., Cano F. R., Lampen J. O. Lomofungin, an inhibitor of ribonucleic acid synthesis in yeast protoplasts: its effect on enzyme formation. Antimicrob Agents Chemother. 1973 Jun;3(6):716–722. doi: 10.1128/aac.3.6.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Cooper T. G. Effects of inducer addition and removal upon the level of allophanate hydrolase in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1973 Dec 19;55(4):1100–1104. doi: 10.1016/s0006-291x(73)80008-7. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Cooper T. G. Kinetics of induced and repressed enzyme synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975 Mar;121(3):1064–1073. doi: 10.1128/jb.121.3.1064-1073.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawther R. P., Phillips S. L., Cooper T. G. Lomofungin inhibition of allophanate hydrolase synthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1975;137(2):89–99. doi: 10.1007/BF00341675. [DOI] [PubMed] [Google Scholar]
- MIDDELHOVEN W. J. THE PATHWAY OF ARGININE BREAKDOWN IN SACCHAROMYCES CEREVISIAE. Biochim Biophys Acta. 1964 Dec 9;93:650–652. doi: 10.1016/0304-4165(64)90349-6. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Cooper T. G. Evidence that specific and "general" control of ornithine carbamoyltransferase production occurs at the level of transcription in Saccharomyces cerevisiae. J Bacteriol. 1977 Jun;130(3):1253–1261. doi: 10.1128/jb.130.3.1253-1261.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Wiame J. -M. The control of ornithinetranscarbamylase activity by arginase in Saccharomyces cerevisiae. FEBS Lett. 1969 Apr;3(1):47–49. doi: 10.1016/0014-5793(69)80093-1. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. Enzyme repression in the arginine pathway of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1969;35(2):215–226. doi: 10.1007/BF02219132. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. Induction and repression of arginase and ornithine transaminase in baker's yeast. Antonie Van Leeuwenhoek. 1970;36(1):1–19. doi: 10.1007/BF02069003. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. The derepression of arginase and of ornithine transaminase in nitrogen-starved baker's yeast. Biochim Biophys Acta. 1968 Mar 11;156(2):440–443. doi: 10.1016/0304-4165(68)90284-5. [DOI] [PubMed] [Google Scholar]
- Penninckx M. Interaction between arginase and L-ornithine carbamoyltransferase in Saccharomyces cerevisiae. The regulatory sites of arginase. Eur J Biochem. 1975 Oct 15;58(2):533–538. doi: 10.1111/j.1432-1033.1975.tb02402.x. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E. Messenger RNA turnover in mouse L cells. J Mol Biol. 1973 Oct 5;79(4):681–696. doi: 10.1016/0022-2836(73)90071-5. [DOI] [PubMed] [Google Scholar]
- Petersen N. S., McLaughlin C. S., Nierlich D. P. Half life of yeast messenger RNA. Nature. 1976 Mar 4;260(5546):70–72. doi: 10.1038/260070a0. [DOI] [PubMed] [Google Scholar]
- Spradling A., Hui H., Penman S. Two very different components of messenger RNA in an insect cell line. Cell. 1975 Feb;4(2):131–137. doi: 10.1016/0092-8674(75)90119-1. [DOI] [PubMed] [Google Scholar]
- Stiles C. D., Lee K. L., Kenney F. T. Differential degradation of messenger RNAs in mammalian cells. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2634–2638. doi: 10.1073/pnas.73.8.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonart P., Bechet J., Hilger F., Burny A. Thermosensitive mutations affecting ribonucleic acid polymerases in Saccharomyces cerevisiae. J Bacteriol. 1976 Jan;125(1):25–32. doi: 10.1128/jb.125.1.25-32.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P., Ramos F., Piérard A., Grenson M., Wiame J. M. Regulation of the carbamoylphosphate synthetase belonging to the arginine biosynthetic pathway of Saccharomyces cerevisiae. J Mol Biol. 1972 Jun 20;67(2):277–287. doi: 10.1016/0022-2836(72)90241-0. [DOI] [PubMed] [Google Scholar]
- Tonnesen T., Friesen J. D. Inhibitors of ribonucleic acid synthesis in Saccharomyces cerevisiae: decay rate of messenger ribonucleic acid. J Bacteriol. 1973 Sep;115(3):889–896. doi: 10.1128/jb.115.3.889-896.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney P. A., Cooper T. G. Requirement for HCO3- by ATP: urea amido-lyase in yeast. Biochem Biophys Res Commun. 1970 Aug 24;40(4):814–819. doi: 10.1016/0006-291x(70)90975-7. [DOI] [PubMed] [Google Scholar]
- Whitney P. A., Cooper T. G. Urea carboxylase and allophanate hydrolase. Two components of adenosine triphosphate:urea amido-lyase in Saccharomyces cerevisiae. J Biol Chem. 1972 Mar 10;247(5):1349–1353. [PubMed] [Google Scholar]
- Whitney P. A., Magasanik B. The induction of arginase in Saccharomyces cerevisiae. J Biol Chem. 1973 Sep 10;248(17):6197–6202. [PubMed] [Google Scholar]
- Wickerham L. J. A Critical Evaluation of the Nitrogen Assimilation Tests Commonly Used in the Classification of Yeasts. J Bacteriol. 1946 Sep;52(3):293–301. [PMC free article] [PubMed] [Google Scholar]



