Abstract
Formulas are derived for the effect of size on a free-swimming microbe’s ability to follow chemical, light, or temperature stimuli or to disperse in random directions. The four main assumptions are as follows: (i) the organisms can be modeled as spheres, (ii) the power available to the organism for swimming is proportional to its volume, (iii) the noise in measuring a signal limits determination of the direction of a stimulus, and (iv) the time available to determine stimulus direction or to swim a straight path is limited by rotational diffusion caused by Brownian motion. In all cases, it is found that there is a sharp size limit below which locomotion has no apparent benefit. This size limit is estimated to most probably be about 0.6 μm diameter and is relatively insensitive to assumed values of the other parameters. A review of existing descriptions of free-floating bacteria reveals that the smallest of 97 motile genera has a mean length of 0.8 μm, whereas 18 of 94 nonmotile genera are smaller. Similar calculations have led to the conclusion that a minimum size also exists for use of pheromones in mate location, although this size limit is about three orders of magnitude larger. In both cases, the application of well-established physical laws and biological generalities has demonstrated that a common feature of animal behavior is of no use to small free-swimming organisms.
Evolution is constrained by the laws of physics and chemistry that describe the world in which organisms live, and it is important to understand how these constraints shape adaptive landscapes and the direction of evolution. Although the physical interactions between most organisms and their environment are complex, the physical forces impacting small organisms that live in water away from surfaces can be described by simple laws. Their environment is uniform in its mechanical properties, flow is laminar, and diffusion controls the distribution of nutrients. This view applies to bacteria in a drop of water as well as micro plankton in larger bodies of water. In such a simple world, it is possible to rigorously determine the circumstances in which locomotion benefits the organism.
Speed
I assume that for typical organisms locomotion is driven by a certain power per unit volume (specific power), which is independent of size. How good is this assumption? Considering all kinds of organisms (with volumes ranging over nearly 20 orders of magnitude), measured specific metabolic rates vary over only 3 orders of magnitude: 0.01–10 ml O2⋅g−1⋅hr−1 (1). Furthermore, the rates do not vary systematically with size—the highest rates are generally found in bacteria and birds, whereas the lowest rates occur in algae and reptiles. Thus, the assumption that available energy is proportional to volume appears to reflect a fundamental tendency of known organisms, and I assume a typical value of 1 ml O2⋅g−1⋅hr−1 (= 5.6 × 104 erg⋅cm−3⋅s−1).
This is the total specific power available to the organism from metabolism, for all functions including inefficiencies. Assuming that 10% of the total energy budget is devoted to locomotion, the specific power available from metabolism to apply to locomotion is 6 × 103 erg⋅cm−3⋅s−1. To determine the power effectively applied to propulsion, the metabolic power must be multiplied by the locomotor efficiency, which is the product of the mechanical efficiency of propulsion, about 10% for bacteria (2), and the efficiency of conversion of electrochemical energy of the proton gradient to torque, about 5% (3). Thus, the overall efficiency is estimated to be about 0.5%, which is consistent with measurements on a rotifer (4) and copepod (5), and this value is assumed here. Consequently, the specific power effectively applied to overcome drag is PV = 30 erg⋅cm−3⋅s−1. (See Table 1 for a summary of symbols used and values assumed.)
Table 1.
Parameters used
| Symbol | Meaning | Value* | Units |
|---|---|---|---|
| α | Light attenuation coefficient | 100 | cm−1 |
| C | Concentration of a chemical | 1 × 1016 | molecules⋅cm−3 |
| d | Distance between positions in a gradient | — | cm |
| D | Diffusion coefficient for chemicals | 1 × 10−5 | cm2⋅s−1 |
| D0 | Diffusion coefficient of a non-motile organism | — | cm2⋅s−1 |
| Dm | Diffusion coefficient of a motile organism | — | cm2⋅s−1 |
| f | Fraction of light absorbed by photoreceptor | 0.0003 | |
| η | Viscosity of the environment | 0.01 | poise = erg⋅s⋅cm−3 |
| Hc | Heat capacity per unit volume | 4.2 × 107 | erg⋅cm−3⋅K−1 |
| HT | Thermal conductivity | 6.2 × 104 | erg⋅s−1⋅cm−1⋅K−1 |
| I | Light intensity | 1 × 1016 | photons⋅s−1⋅cm−2 |
| k | Boltzmann’s constant | 1.4 × 10−16 | erg⋅K−1 |
| L | Decay length of spatial gradient | —† | cm |
| PV | Specific mechanical power | 30 | erg⋅cm−3⋅s−1 |
| r | Radius of organism | — | cm |
| T | Absolute temperature | 293 | °K |
| u | Speed relative to size | 20 | radii⋅s−1 |
| 10 | diameters⋅s−1 | ||
| v | Speed of swimming | — | cm⋅s−1 |
Indicates the specific values assumed in making the standard estimates of Table 2.
A value of 0.1 cm for chemical and light stimuli; 5 × 105 cm for temperature.
Assuming that organisms of interest can be approximated by a sphere of some radius, r, the speed of swimming (v) can be calculated from Stokes’ Law, and the speed is (6) v = r(2 PV/9 η)½, where η is the viscosity of the medium, which is assumed to have the value 0.01 poise (appropriate for water, under normal conditions). From this equation, the absolute speed (v) is proportional to the radius (or diameter), and the relative speed (u = v/r) is predicted to be independent of size. With the assumed value of PV, the predicted speed is close to 13 diameters/s. Because of the square-root relationship, any change in the assumed values of energy allocation or efficiency have a relatively small impact on the speed.
Stokes’ law is valid when the Reynolds’ number is less than 0.5 (7). This criterion is satisfied in water when u = 10 diameters/s and diameter is less than 220 μm. Even for the highest speeds of 100 diameters/s, it is satisfied for sizes as large as 70 μm diameter. These limits are far larger than the sizes we will be concerned with, so speed is well within the range where Stokes’ law applies.
Observations of the highest sustained speeds for all kinds of swimming organisms, from bacteria to whales, indicates that relative speed is comparatively uniform (8). Although speeds and lengths vary more than a million-fold, relative speeds vary only about a hundred-fold—from 1 to 100 lengths/s, with a most common value near 10 lengths/s. These observations agree with the previous calculation, and a value of 10 diameters (20 radii)/s is assumed below.
Dispersal
All organisms face the problem of dispersing progeny away from one another and to new environments. Free-floating cells of micrometer size will be moved by Brownian motion, and their behavior can be simply described by a diffusion coefficient. For such nonmotile organisms, the diffusion coefficient is (ref. 9, p. 56) D0 = k T/6 π η r, where k is Boltzmann’s constant (1.38 × 10−16 erg⋅K−1), and T is absolute temperature (20°C = 293°K). If the organism swims, it can move faster, and the straighter its path, the further it will disperse in a given time. If the organism does not use a so-called collimating stimulus (10) to guide its locomotion, the best it can do is to swim as straight as it can, but this will be limited by random rotations caused by Brownian motion. In this case, the behavior can be described by a modified diffusion coefficient. Berg (ref. 9, p. 94) has found that the diffusion coefficient for a free-swimming organism is Dm = v2/6 Dr, where v is the absolute speed and Dr is the rotational diffusion coefficient for the organism, which is k T/8 π η r3 (refs. 9, p. 83; 11, p. 435). Substituting this formula for Dr and u r for v, I find that the effective diffusion coefficient for a motile organism is Dm = 4 π η u2 r5/3 k T. To measure the effect of motility on dispersal, take the ratio of the diffusion coefficients with and without motility to obtain the first formula in Table 2. In order for motility to double the diffusion coefficient for a motile organism (Dm = 2 D0) the organism must have a diameter of at least 0.64 μm, using the standard parameter values (Table 1). Because the ratio of the diffusion coefficients varies as the sixth power of the radius, the size limit is sharply defined and is relatively insensitive to the values of the other parameters.
Table 2.
Constraints on size
| Stimulus | Mechanism | Constraint formulas | 2 r, μm |
|---|---|---|---|
| None | Diffusion | 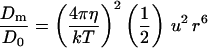 |
0.64 |
| Chemical | Spatial | 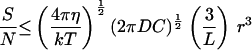 |
0.58 |
| Chemical | Temporal | 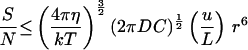 |
0.65 |
| Light | Spatial | 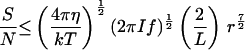 |
1.77 |
| Light | Temporal | 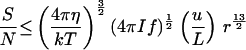 |
1.05 |
| Light | Direction | 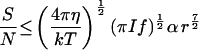 |
1.24 |
| Temperature | Spatial | 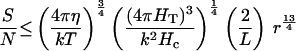 |
0.74 |
| Temperature | Temporal | 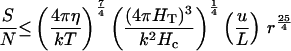 |
0.69 |
In the constraint formulas, the parameters are grouped so that the term on the left is a performance factor (signal-to-noise ratio or ratio of diffusion coefficients with and without motility), the first term to the right of the equal sign involves hydrodynamics, the second term involves stimulus intensity, the third term involves gradient magnitude and/or relative speed, and the right-most term involves the size of the organism. The right-hand column gives the minimum diameter an organism can have and usefully employ the indicated mechanism, assuming the parameter values in Table 1 and S/N = (Dm − D0)/D0 ≥ 1 for useful function.
Nutrient Uptake
Motile microbes must obtain nutrients by the diffusion of individual molecules to the surface of the organism. Can this process be enhanced by swimming? Although there is broad agreement that this can occur only for larger organisms, this question has proven to be difficult to analyze quantitatively, and two points of view (12, 13) have yet to be reconciled. Fortunately, both analyses lead to the same conclusion for the conditions of interest here. Making the most optimistic assumption that metabolic energy is proportional to the rate of nutrient uptake, the assumption that 10% of the organism’s energy metabolism is devoted to locomotion (to attain speeds of 10 diameters/s), requires a 10% increase in nutrient uptake to pay for its cost. Using the values in Table 1, the numerical analysis of Berg and Purcell (12) indicates that to meet this requirement a minimum diameter of 3.7 μm is necessary, whereas the relation favored by Karp-Boss et al. (13) predicts a minimum diameter of 8.5 μm. In either case, it is clear that the smallest motile bacteria are too small to benefit from motility by enhanced nutrient uptake. The benefit of moving away from one’s own waste products should have a similar size dependence.
Orientation by Stimuli
Another potential benefit of locomotion is that it may be oriented by light direction or along chemical, light, or temperature gradients to move an organism to a more favorable environment or (used as collimating stimuli) to keep it moving in one direction to increase dispersal or the efficiency of searching (ref. 14, pp. 396–397).
Gradients.
In a common environmental situation, a certain concentration of nutrient is maintained constant in one layer by mixing and it diffuses into a stationary layer where it is consumed at a rate proportional to its local concentration; the constant of proportionality is equal to the time constant, τ, for decline of local concentration if the supply is stopped. In this situation, the concentration declines exponentially with distance, x, from the boundary [C = C0 exp(−x/L)], with a decay length of L = 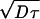 , where D is the diffusion constant of the nutrient.
, where D is the diffusion constant of the nutrient.
Directing locomotion along a gradient requires that an organism measure intensity (concentration, temperature, or light intensity) sufficiently accurately at two different positions in the gradient, which I assume are separated by a distance d. For determining the direction of a gradient, the signal (S) is the difference in intensity (ΔI) between the two locations. For the exponential gradient, ΔI = Im [1 − exp(−d/L)], where Im is the intensity at the location and L is the gradient decay length. In cases of interest here, d ≪ L, and the exponential function can be accurately approximated by the first terms of its series representation. This gives the signal S = ΔI ≅ Im d/L.
The gradient decay length, L, is also equal to the reciprocal of the relative gradient, (1/I) dI/dx, which can be used to characterize the local steepness of any gradient that varies smoothly with little change in intensity over the distance d, as holds for cases of interest here. For any such gradient, S ≅ Ieff d/L, where Ieff is the effective intensity at the location, and L will be used as the measure of gradient steepness.
Estimates of the order of magnitude of L for chemical gradients can be obtained from measurements of oxygen, sulfate, and ammonia in the pore water of sediments at the bottom of aerated fresh and salt waters (15, 16). Observed decay lengths varied from 0.01 to 1 cm, and I generally assume a value of L = 0.1 cm. [In the lab, bacteria have been observed to move up gradients with decay lengths of 0.4 to 5 cm (17).] Concentrations varied from 5 to 350 μM or 3 × 1015 to 2 × 1017 molecules/cm3, and I generally assume a value of C = 1 × 1016 molecules/cm3 (= 17 μM).
For nutrients and chemical stimuli, the diffusion coefficient, D, is assumed to have a value of 1 × 10−5 cm2⋅s−1. This is about the value for glycine in water, whereas oxygen and carbon dioxide have diffusion coefficients about twice as large and sugars about half as large.
Relevant light intensity gradients are probably those at the edge of shadows or in sediments and are likely of similar magnitude to the chemical gradients (L = 0.1 cm).
The cooling of the earth produces an average ground temperature gradient of 6 × 10−4 °C/cm (18). Temperature gradients in the top 100 m of open ocean waters are frequently of the same order of magnitude (19). At 20°C, a gradient of this magnitude is equivalent to a decay length of L = [(1/T) dT/dx]−1 = [(1/293°K) × 6 × 10−4 °K/cm]−1 = 5 × 105 cm, and this value will be assumed for temperature. (This value is large because natural temperature differences on the earth’s surface are small fractions of absolute temperature, and the decay length measures the distance necessary to get to absolute zero.)
Orientation.
There are two basic mechanisms free-swimming organisms can use to determine gradient direction (ref. 14, pp. 414–416). Those making spatial comparisons simultaneously compare the intensity of stimulation of receptors in different parts of the organism, which allows the organism to turn in the appropriate direction (tropotaxis). In this case, d ≤ 2 r. Organisms making temporal comparisons, compare the intensity of stimulation of receptors at different times, between which the organism moves from one location (or orientation in the case of light direction) to another, and modulate the probability of changing their direction of locomotion (klinokinesis or klinotaxis). In this case, d ≤ v t = u r t, where t is the time between measurements.
For free-swimming microbes, the time available to make measurements is limited by the rate of random rotation caused by Brownian motion. The comparison with the past provides no useful information if past positions (or orientations) are equally likely to be in any direction from the present position (or orientation). To estimate the useful time interval available between measurements, consider the average over a population of organisms of the cosines of angles changed from an original orientation; this average decays exponentially from one to zero with a time constant of (ref. 11, pp. 432–437)
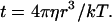 |
1 |
I will use this time constant for an estimate of the maximum interval over which intensity and gradient direction can usefully be measured.
Another constraint on the organism’s ability to detect a gradient, is the noise (N) in measuring the signal. In general, noise can be reduced by averaging the signal over larger sizes and longer time periods, but time is limited and noise is reduced only as the square root of time or size. For significant reliability, the signal-to-noise ratio must be greater than about one, (ref. 14, p. 94) and I assume S/N = 1, at the limit of useful function.
Chemical Stimuli.
It is assumed that the organism has the optimal design of numerous receptors covering the whole surface and that binding saturates only at higher concentrations than present in the environment. The analysis of spatial comparison presented here will assume that the comparison is between one half sphere and the other. Berg and Purcell (12) have considered the case of fore–aft comparison in chemical gradients and found a significant complication in that the movement of the organism increases diffusion to the leading surface and reduces it to the trailing surface. This complicates detection of an actual gradient, but in principle the organism might be able to correct for this effect, and I will ignore it. Suggestions that Escherichia coli might take advantage of the increased flux to its leading edge have been discounted by further observations (20). Although no unattached bacteria are known to employ spatial comparison, most organisms that do, use a lateral distribution of receptors rather than fore-and-aft (ref. 14, p. 430), and this mechanism should not suffer from unequal diffusion.
For spatial comparison detecting a chemical gradient, I used a formula for the diffusive current to half a stationary sphere (ref. 12, equation D3, corrected according to ref. 21). The currents (J) to the half on the high concentration side of the sphere (+) and to the low-concentration side (−) are given by J± = 2 π D C r (1 ± 3 r/2 L). The signal is taken as the difference in the number of molecules arriving at the surface between the two sides in a time, t or (J+ − J−) t, which gives a signal S = 6 π D C t r2/L. The noise is the standard deviation of the mean count, so N = (2 π D C r t)½. The signal-to-noise ratio is then, S/N = 3 (2 π D C t)½ r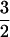 /L. Substituting Eq. 1 for t gives the second equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 0.58 μm diameter to usefully orient to chemical gradients by spatial comparisons.
/L. Substituting Eq. 1 for t gives the second equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 0.58 μm diameter to usefully orient to chemical gradients by spatial comparisons.
For temporal detection of a chemical gradient, Ieff = C, and N = C (2 π D C r t)−½ (ref. 12, equation. 55). The signal-to-noise ratio is then, S/N = (2 π D C r t)½ d/L. Substituting Eq. 1 for t gives the third equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 0.65 μm diameter to usefully orient to a chemical gradients by temporal comparisons.
Light.
For gradients of light, Ieff = I A f t, where I is the number of photons passing a unit area in unit time, A is the area of the photoreceptor, and f is the fraction of photons striking the receptor area that is absorbed (which is generally much less than one). The assumed values of I (1 × 1016 photons⋅s−1⋅cm−2) and f (0.0003) are for full sunlight at the earth’s surface captured by a single membrane packed with rhodopsin molecules (ref. 14, pp. 161, 166, 178). The noise is the standard deviation of the photon count. Because photon counts follow a Poisson distribution, noise is simply the square root of the mean, and N = (I A f t)½.
For spatial comparisons, A ≤ 2 π r2, half the surface area of the organism. The signal-to-noise ratio is S/N = (I A f t)½ r/L. Substituting for A and Eq. 1 for t gives the fourth equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 1.77 μm diameter to usefully orient to light gradients by spatial comparisons.
For temporal comparisons, A ≤ 4 π r2, the whole surface area of the organism. Assuming that the distance moved before orientation is lost is small compared with the light gradient decay length, the signal can be approximated as I A f t u r t/L and the signal-to-noise ratio is S/N = (I A f t)½ u r t/L. Strictly speaking, the two t’s are different: the first is the time period over which a measurement is made, whereas the second is the time between measurements. However, maximum performance requires that the first be as large as possible, which is of the same order as the second time interval. Consequently I estimate both of them by the rotational diffusion time (Eq. 1) and obtain the fifth equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 1.05 μm diameter to usefully orient to a light gradients by temporal comparisons.
Detection of the direction of propagation of light requires significant attenuation of light traversing the organism, depending on the attenuation coefficient (α) and thickness of the optical screen (x). It is assumed that α = 100 cm−1, which is close to the value found in the photoreceptor cells of animals (ref. 14, p. 168). For sizes of interest here, α x ≪ 1, and Lambert’s Law can be approximated by a linear relationship, giving S ≅ I A f t α x, and S/N = (I A f t)½ α x. To fit within a sphere, x ≤ 2 r and A ≤ π r2 (the cross-sectional area of the sphere), but both cannot have these maximal values simultaneously. To estimate an optimal arrangement, I assume that the screen is a cylinder enclosed within the spherical organism with its axis (of length x) parallel to the direction of light propagation at the optimal orientation of the organism and that the receptor area is equal to a flat end of the cylinder. (Spherical ends only increase the area about 10% and lead to a more complicated formula.) S/N is proportional to x 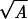 , which has a maximal value of
, which has a maximal value of 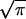 r2, under these assumptions. Using this and Eq. 1 for t gives the sixth equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 1.24 μm diameter to usefully orient to light direction.
r2, under these assumptions. Using this and Eq. 1 for t gives the sixth equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 1.24 μm diameter to usefully orient to light direction.
In reality, most phototactic cells employ a single receptor that is rotated around the axis of locomotion and scans the environment like a radar (22, 23). However, this mechanism faces the same basic constraints in obtaining a signal to orient to the light, and the size limit should be the same as for the simultaneous detection of light direction calculated above.
Temperature.
For temperature gradients, Ieff = T, the absolute temperature, and the signal is S = T d/L. The temperature fluctuation noise experienced by a cell less than 300 μm in extent when averaged over a time period t is (18) N = T (k/Hc)½ (Hc/4 π HT t)¾, where Hc is the heat capacity per unit volume (4.2 × 107 erg⋅cm−3⋅K−1 for water), and HT is the thermal conductivity of the organism and its immediate environment (6.2 × 104 erg⋅s−1⋅cm−1⋅K−1 for water).
The signal-to-noise ratio is then
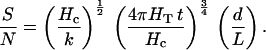 |
2 |
For the time period, t, I again use Eq. 1. For a spatial comparison, d ≤ 2 r, giving the seventh equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 0.74 μm diameter to usefully orient to a temperature gradient by spatial comparisons.
For a temporal comparison, d is at best v t = u r t, and I obtain the last equation in Table 2. With the estimated parameter values (Table 1), an organism must be larger than 0.69 μm diameter to usefully orient to a temperature gradient by temporal comparisons.
Bacteria that contain magnets that force the organism into alignment with the earth’s magnetic field (24) have not been treated because they are not free, but even more constrained in their locomotor movements than organisms moving in contact with two-dimensional surfaces.
Prediction
In all these cases, there exists an absolute size limit below which directed movement is impossible for free-swimming organisms—no matter which mechanism they employ. Remarkably, in all cases, performance is proportional to large powers of linear size, and the actual values assumed for the other parameters influence the size limits only weakly. Using the most favorable plausible values for parameters characterizing the organism and typical values for parameters characterizing the environment, the limits fall within the narrow range of 0.6 to 1.8 μm (Table 2).
Because no other benefits of swimming are evident and the size limits are insensitive to the assumed values of the parameters (because of the much larger powers of size), I conclude that a free-floating organism smaller than 0.6 μm diameter is unlikely to obtain any advantage by expending energy on swimming. Applying these conclusions for spherical organisms to other shapes, I simply equate the diameter of the sphere to the largest dimension (length), because that is the dimension that provides the slowest rotational diffusion. Thus, I arrive at the prediction that the smallest free-swimming bacteria have a length greater than 0.6 μm.
There has previously been considerable speculation about the smallest possible bacterium (25). Theoretical considerations suggest limits of 0.1–0.2 μm diameter, and the smallest well-documented bacteria are about this size. Because this is several-fold smaller that the predicted minimum size for useful motility, it is predicted that there can exist a group of small nonmotile bacteria, with lengths in the range of 0.1–0.6 μm.
Observation
To test these predictions, I checked each genus of bacteria described in all four volumes of Bergey’s Manual of Systematic Bacteriology (26). The goal was to collect an unbiased sample of actual microbes, and objective criteria were established for the selection of data. Only genera that appeared to consist of unattached, free-swimming or free-floating types were included; genera described as having mycelial growth forms, gliding motility, containing magnetic particles, or engaging in intracellular parasitism were excluded. In most cases, a numerical range for both width and length was given, and the geometric mean (= arithmetic mean on the log scale) of the length range was used for the size parameter.
The motility status for each genus was recorded as (i) nearly all strains (or individuals) motile, (ii) nearly all nonmotile, or (iii) mixed (some motile while others were not). Only one set of size range and motility status was recorded for each genus in the hope of reducing the over-representation of human pathogens that would have occurred if each described species contributed a data set. If size was not described numerically or motility status was not indicated for the genus, the type species for the genus was examined, and if it’s description was deficient, the first species description in the genus that contained the required information was used. When a genus was described in more than one location, only the first description was used. This procedure produced usable data for 218 genera, of which 97 were characterized as motile, 94 as nonmotile, and 27 as mixed.
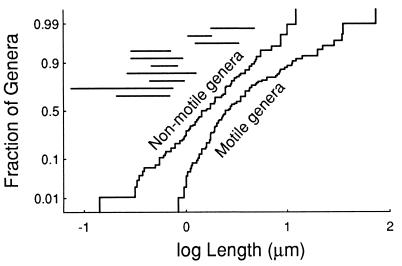
To compare the motile and nonmotile genera, I analyzed distributions of the logarithms of lengths. Parametric analysis of the frequency distribution indicated that the group of 94 nonmotile genera fit a normal distribution (skewness = 0.002, kurtosis = 0.033), whereas the 97 motile genera did not (skewness = 1.165, kurtosis = 1.155). This is seen in a probability plot (27) of the cumulative frequency distributions (Fig. 1). In this plot, the distribution of nonmotile genera falls close to a straight line (as expected for a normal distribution), whereas the motile distribution is clearly curved, with fewer small genera and more larger ones than expected. Nonparametric statistical tests (Mann–Whitney U and Kolmogorov–Smirnov) indicate that the probability that the two groups are from the same size distribution is less than 10−4.
Figure 1.
Cumulative frequency plots of length distributions for motile and nonmotile genera. The stepped curves represent the fraction of genera for which the length is less than the value on the horizontal axis. (Where a range of lengths was reported, the geometric mean of the range is used, which is the midpoint on the log scale.) The vertical axis is a normal probability scale (27), which causes a cumulative normal distribution to fall on a straight line. The motile genera range in size from 0.84 to 73 μm, whereas the nonmotile genera range over 0.14 to 12 μm. Note that the small size limit of the motile group is cutoff more sharply than a normal distribution would be and remains above the predicted size limit of about 0.6 μm. In addition, the motile group has more large genera than expected for a normal distribution. The horizontal lines represent the range of predicted size limits when parameters range over the limits used in the Discussion. From the top down, they are for light stimuli with (spatial, temporal, and directional orientation mechanisms), followed by dispersal, then chemical concentration (spatial and temporal), chemical gradient steepness (spatial and temporal), and finally temperature (spatial and temporal).
Examination of the distributions reveals that 8% (8/97) of the motile genera are larger than the largest nonmotile genus, and, more to the point, 19% (18/94) of the nonmotile genera are smaller than the smallest motile genus, which has a length of 0.8 μm. In addition, 10 of the nonmotile genera are smaller than the predicted 0.6-μm limit, whereas none of the motile genera are this small. The prediction that a lower size limit for motility exists was tested by calculating (from the hypergeometric distribution) the probability that the 18 smallest genera would all be nonmotile (as observed), if motility were distributed independently of size, obtaining P < 2 × 10−6. The prediction that the size limit is 0.6 μm was tested similarly for the 10 genera smaller than 0.6 μm, obtaining P < 7 × 10−4.
Discussion
These observations confirm the prediction that there is a lower size limit to useful motility by free-floating organisms. The actual value of this limit depends on the values of the parameters, most importantly parameters describing effective stimulus intensity and gradient steepness. The standard values were chosen because they were well-defined values for some common natural situations. However, it should be recognized that other values may occur in other situations and give rise to somewhat different size limits.
A review of metabolic rates (28) suggests that 95% of bacteria have metabolic rates in the range 0.35 to 74 ml O2·g−1⋅h−1, which corresponds to 2 × 104 to 4 × 106 erg⋅cm−3⋅s−1. With the assumptions previously made for energy allocation and efficiencies, this range of metabolic rates would suggest relative speeds in the range 7.5–105 diameters/s. With the standard values (Tables 1 and 2) for the other parameters, this range of speeds indicates size limits for dispersal of 0.7–0.3 μm diameter.
Similarly, a range of chemical concentrations from 1 μM to 1 mM gives limiting diameters of 0.9–0.3 μm (spatial comparison mechanism) and 0.8–0.5 μm (temporal). Concentration gradients producing decay lengths in the range 0.01–1 cm−1 give limiting diameters of 0.3–1.2 μm (spatial) and 0.4–1.0 μm (temporal).
Light intensity for organisms does not get brighter that sunlight, although an organism might capture a somewhat higher fraction of photons than assumed. On the other hand, there is no practical limit to how dim light can be. A guess as to the minimum light intensity useful to bacteria of 0.1% of full sunlight (1013 photons⋅cm−2⋅s−1 in the wavelength range of absorption) leads to predicted limiting diameters of 4.8 μm (spatial), 1.8 μm (temporal), and 3.3 μm (direction).
Although thermotaxis is rarely reported for bacteria, temperature gradients provide the lowest plausible size limits. Temperature gradients easily reach 1°C/cm where sunlight shines directly on soil (18). Gradients of this magnitude have decay lengths L = [(1/T) dT/dx]−1 = [(1/293°K) × 1°K/cm]−1 = 293 cm. This is larger than the previously assumed value (Table 1) by a factor of 1,700. The last two equations of Table 2 indicate that organisms could orient to such steep gradients if they had diameters as small as 0.2 μm (temporal) or 0.07 μm (spatial). Thus, in steep thermal gradients, the size limit approaches the size of the smallest bacteria.
It has often been stated that bacteria are too small to make practical use of spatial comparisons. However, the relations of Table 2 demonstrate that the size limits for spatial and temporal comparisons are impacted by different powers of the parameters. Consequently, there are some situations in which an organism using spatial comparisons can be smaller than one using the more common temporal comparison. This issue will be treated more thoroughly in another paper.
There are certainly other possible explanations for the observed size limit on motility. For example, it could be that the only motors that have evolved have a minimum size or a minimum power requirement. However, the explanation proposed here is more fundamental. It says that there is no use for motility in smaller organisms because of constraints imposed by physical laws, and thus motility can never evolve in these organisms.
Considerations similar to those used in this paper have previously led to the conclusion that there exists a minimum size for use of pheromones in mate location, although this size limit is about 3 orders of magnitude larger (6). In both cases, the rigorous application of well-established physical laws and biological generalities has demonstrated that common features of animal behavior are of no use to small free-swimming organisms.
Acknowledgments
I thank Edward Yeargers, Ronald Fox, Terry Snell, Mark Borodovsky, Howard Berg, and four anonymous reviewers for making valuable suggestions on previous drafts of the manuscript.
References
- 1.Altman P L, Dittmer D S. Biology Data Book. Bethesda, MD: Fed. Am. Soc. Exp. Biol.; 1974. pp. 1613–1659. [Google Scholar]
- 2.Yates G T. Am Sci. 1986;74:358–365. [Google Scholar]
- 3.Meister M, Lowe G, Berg H C. Cell. 1987;49:643–650. doi: 10.1016/0092-8674(87)90540-x. [DOI] [PubMed] [Google Scholar]
- 4.Epp R W, Lewis W M., Jr Evolution. 1979;33:919–928. doi: 10.1111/j.1558-5646.1979.tb04745.x. [DOI] [PubMed] [Google Scholar]
- 5.Vlymen W. Limnol Oceanogr. 1970;15:348–356. [Google Scholar]
- 6.Dusenbery D B, Snell T W. J Chem Ecol. 1995;21:427–438. doi: 10.1007/BF02036740. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson G E. A Treatise on Limnology. Vol. 2. New York: Wiley; 1967. pp. 258–259. [Google Scholar]
- 8.Dusenbery D B. Life At Small Scale: The Behavior of Microbes. New York: Scientific American Library; 1996. p. 45. [Google Scholar]
- 9.Berg H C. Random Walks in Biology. Princeton: Princeton Univ. Press; 1993. [Google Scholar]
- 10.Pline M, Dusenbery D B. J Chem Ecol. 1987;13:873–888. doi: 10.1007/BF01020167. [DOI] [PubMed] [Google Scholar]
- 11.Tanford C. Physical Chemistry of Macromolecules. New York: Wiley; 1963. pp. 432–437. [Google Scholar]
- 12.Berg H C, Purcell E M. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karp-Boss L, Boss E, Jumars P A. Oceanogr Mar Biol. 1996;34:71–107. [Google Scholar]
- 14.Dusenbery D B. Sensory Ecology. New York: Freeman; 1992. [Google Scholar]
- 15.Canfield D E, Jørgensen B B, Fossing H, Glud R, Gundersen J, Ramsing N B, Thamdrup B, Hansen J W, Nielsen L P, Hall P O J. Mar Geol. 1993;113:27–40. doi: 10.1016/0025-3227(93)90147-n. [DOI] [PubMed] [Google Scholar]
- 16.Ford T E. Aquatic Microbiology: An Ecological Approach. Boston: Blackwell; 1993. p. 84. [Google Scholar]
- 17.Dahlquist F W, Elwell R A, Lovely P S J. Supramol Struct. 1976;4:329–342. doi: 10.1002/jss.400040304. [DOI] [PubMed] [Google Scholar]
- 18.Dusenbery D B. J Theor Biol. 1988;131:263–271. doi: 10.1016/s0022-5193(88)80224-8. [DOI] [PubMed] [Google Scholar]
- 19.Urick R J. Principles of Underwater Sound. New York: McGraw Hill; 1983. , Figs. 5.13 and 5.17. [Google Scholar]
- 20.Berg H C, Turner L. Proc Natl Acad Sci USA. 1995;92:477–479. doi: 10.1073/pnas.92.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg H C, Purcell E M. In: Biological Physics. Mielczarek E V, Greenbaum E, Knox R S, editors. New York: American Institute of Physics; 1993. p. 123. [Google Scholar]
- 22.Foster K W, Smyth R D. Microbiol Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crenshaw H C. Am Zool. 1996;36:608–618. [Google Scholar]
- 24.Blakemore R P. Annu Rev Microbiol. 1982;36:217–238. doi: 10.1146/annurev.mi.36.100182.001245. [DOI] [PubMed] [Google Scholar]
- 25.Various. Science. 1997;276:1776–1777. [PubMed] [Google Scholar]
- 26.Holt, J. G. (1984, 1986, 1989) Bergey’s Manual of Systematic Bacteriology (Williams & Wilkins, Baltimore), Volumes 1–4.
- 27.Sokal R R, Rohlf F J. Biometry. San Francisco: Freeman; 1981. p. 118. [Google Scholar]
- 28.Robinson W R, Peters R H, Zimmermann J. Can J Zool. 1983;61:281–288. [Google Scholar]