Abstract
A better understanding of Mycobacterium tuberculosis virulence mechanisms is highly dependent on the design of efficient mutagenesis systems. A system enabling the positive selection of insertional mutants having lost the delivery vector was developed. It uses ts-sacB vectors, which combine the counterselective properties of the sacB gene and a mycobacterial thermosensitive origin of replication and can therefore be efficiently counterselected on sucrose at 39°C. This methodology allowed the construction of M. tuberculosis transposition mutant libraries. Greater than 106 mutants were obtained, far exceeding the number theoretically required to obtain at least one insertion in every nonessential gene. This system is also efficient for gene exchange mutagenesis as demonstrated with the purC gene: 100% of the selected clones were allelic exchange mutants. Therefore, a single, simple methodology has enabled us to develop powerful mutagenesis systems, the lack of which was a major obstacle to the genetic characterization of M. tuberculosis.
Mycobacterium tuberculosis, the etiologic agent of tuberculosis (TB), infects one-third of the world’s population and kills 3 million people each year. TB is the largest cause of death in the world caused by a single infectious organism (1). According to the World Health Organization, more people died from TB in 1995 than in any other year in history. It has been estimated that, at current rates, up to one-half of a billion people will suffer from TB in the next 50 years. However, despite its importance, the genetic determinants of M. tuberculosis virulence remain poorly characterized. The extreme difficulty in creating defined mutants of M. tuberculosis, either by allelic exchange or transposon mutagenesis, has prevented identification of its virulence factors following Koch’s molecular postulates (2, 3). Rather, alternative genetic strategies have been used, including complementation of nonpathogenic bacteria (4) or spontaneous avirulent mutants with libraries of virulent M. tuberculosis (5) or M. bovis (6) chromosomal DNA. Although these studies have identified genes required for entry into epithelial cells and conferring a growth advantage in vivo, the great majority of the mycobacterial genes involved in virulence remains unknown. Developing efficient mutagenesis systems is thus a top priority for mycobacterial genetics.
Transposon mutagenesis, which has often been used for the identification of bacterial virulence factors, is one of the most powerful methods of mutagenesis. It involves using a mobile element to disrupt genes randomly in the chromosome upon transposition. Using a conventional strategy, i.e., delivering the transposon on a vector unable to replicate in mycobacteria, transposition has been demonstrated in M. bovis Bacille Calmette–Guérin (BCG), a member of the M. tuberculosis complex, for several mycobacterial transposons: IS1096 (7), IS6120, and IS6100 derivatives (unpublished data). Because electroporation efficiencies and transposition frequencies are very low, no more than 100 mutants per experiment have been obtained (7). However, M. tuberculosis contains an estimated 3000 genes, so ≈10,000 mutants would be required to obtain a mutation in each of them (8). Suicide delivery vectors are thus not suitable for the construction of representative libraries of mutants. Another method for creating mutants is allelic exchange mutagenesis. Recently, low frequency allelic exchange was demonstrated in bacteria of the M. tuberculosis complex, again using a suicide delivery vector (9, 10), and new protocols allowing easier detection of allelic exchange mutants also have been developed (11–13). However, as for transposon mutagenesis, detection of very rare allelic exchange events is hindered by low transformation efficiencies and high frequencies of illegitimate recombination. Thus, many genes still remain refractory to allelic exchange by available technology. Clearly, both mutagenesis systems require the design of more efficient methods. The encountered problems can be circumvented using a replicative delivery vector that is efficiently lost under certain conditions. Allowing the introduced delivery vector to replicate avoids the problems arising from low transformation efficiencies. Then, under counterselective conditions, clones that still contain the vector are eliminated, allowing the detection of very rare genetic events. One such system has been developed: Using a conditionally replicative vector that is efficiently lost at 39°C in M. smegmatis, the first mycobacterial insertional mutant libraries were constructed in this fast-growing model strain (14). However, the thermosensitive vectors used are only weakly thermosensitive in slow-growing mycobacteria of the M. tuberculosis complex and therefore cannot be used in these species for transposon or allelic exchange mutagenesis.
Here we describe a system that uses the counterselective properties of the sacB gene and a mycobacterial thermosensitive origin of replication and enables the positive selection of insertion mutants. This methodology was first used to deliver derivatives of IS1096 (15) into the chromosome of M. tuberculosis, leading to the construction of the first transposition mutants libraries for this important pathogen. The libraries were analyzed and seem to be representative. In addition, this method also allowed efficient gene exchange mutagenesis as demonstrated with the purC gene (16): 100% of the selected clones corresponded to allelic exchange mutants.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions.
Escherichia coli DH5α, the strain used in this study for cloning experiments, was routinely grown on liquid or solid Luria–Bertani medium. M. smegmatis mc2155 (17), M. tuberculosis 103 (isolated from a TB patient), and M. bovis BCG Pasteur were grown on liquid Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol and 0.05% Tween or on solid Middlebrook 7H10 medium (Difco). When required, antibiotics were included at the following concentrations: kanamycin (20 μg⋅ml−1) and gentamicin (5 μg⋅ml−1) for mycobacteria and gentamicin (20 μg⋅ml−1) for E. coli. Where indicated, 10 or 2% sucrose was added for M. smegmatis or bacteria of the M. tuberculosis complex, respectively (13, 18).
Purine auxotrophs were identified by their inability to grow on Sauton medium, unless the medium was supplemented with hypoxanthin (20 μg⋅ml−1). In brief, single colonies were picked and resuspended in 96-well microtiter plates containing Sauton medium with or without hypoxanthin supplement. The plates were incubated at 37°C under 5% CO2. Growth was estimated by following the opacity in adjacent wells with and without hypoxanthin addition.
Electrotransformation.
Electrocompetent cells were prepared as described (19) with minor modifications. M. tuberculosis and M. bovis BCG were grown in 200 ml of 7H9 medium to an OD600 of 0.4. Cells were washed three times in 10% glycerol and resuspended in 1 ml 10% glycerol. Aliquots (100 μl) of freshly prepared competent cells were electroporated in the presence of 1 μg of vector DNA in 0.2-cm cuvettes (Bio-Rad) with a single pulse (2.5 kV; 25 μF; 200 ohms). Five milliliters of fresh medium was then added, and the culture was incubated at 32°C for 24 h before plating to allow antibiotic resistance expression. Transformants were scored after 7–8 weeks of incubation at 32°C.
DNA Extraction and Southern Analysis.
Mycobacterial genomic DNA was isolated as described (19) with minor modifications. One hundred microliters of d-cycloserine (mg⋅ml−1) was added to a 10-ml saturated culture that was then incubated overnight at 37°C. Cells were pelleted by centrifugation (15 min at 5000 × g). The pellet was resuspended in 250 μl of solution I (25% sucrose/50 mM Tris⋅HCl, pH 8.0/50 mM EDTA/500 μg⋅ml−1 lysozyme) and incubated overnight at 37°C. Two hundred and fifty microliters of solution II (100 mM Tris⋅HCl, pH 8.0/1% SDS/400 μg⋅ml−1 Proteinase K) was then added, and the samples were incubated for 4 h at 55°C. The lysate was then extracted twice with phenol-chloroform, and the DNA was concentrated by ethanol precipitation. Approximately 1 microgram of genomic DNA was digested overnight with an excess of restriction enzyme (30 units), and the fragments were separated by electrophoresis through 0.7% agarose gels. Southern blotting was carried out in 20× SSPE (150 mM NaCl/8.8 mM NaH2PO4/1 mM EDTA, pH 7.4) using Hybond-N+ nylon membranes (Amersham). The Megaprime random-primed labeling kit (Amersham) and 5 μCi of [α-32P]dCTP were used to label probes. Nonincorporated label was removed by filtration through a Nick Column (Pharmacia). Prehybridization and hybridization were carried out at 65°C using rapid hybridization buffer (Amersham) as recommended by the manufacturer. Serial 15-min washes were performed at 65°C as follows: two washes with 2× SSPE/SDS 0.1%, one wash with 1× SSPE/SDS 0.1%, and two washes with 0.7× SSPE/SDS 0.1%. BioMax MS x-ray film (Kodak) was exposed for 4 h to the blots at −80°C.
Construction of Genomic Libraries.
Chromosomal DNA from transposition mutants was digested with KpnI or BamHI, which do not cut in the transposon, and purified with QIAquick Nucleotide Removal Kit (QIAGEN). A plasmid library was constructed using KpnI- or BamHI-digested and dephosphorylated “ready to clone” pUC18 vectors (Appligene, Strasbourg, France). Transformants corresponding to integrated transposon and flanking sequences were selected on l-kanamycin.
DNA Sequencing.
Sequences of double-stranded plasmid DNA were determined using a DNA Analysis System model 373 stretch (Applied Biosystems) and the Taq Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems). Outward primers based on IS1096 sequence (15) used were: α 5′-CTTCCGCTTCTTCTCCGG-3′ and β 5′-CCATCATCGGAAGACCTC-3′.
Construction of Vectors.
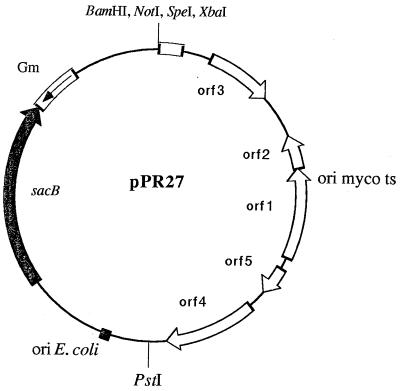
The thermosensitive origin of replication of pAL5000, present in ts-sacB delivery vectors, was extracted from pB4D* on a 5-kb BamHI (whole pAL5000) or a 3.7-kb EcoRV+KpnI (minimal origin of replication) fragment (20). The fragments were blunt-ended and cloned into SmaI-cut pJQ200 harboring the sacB gene (21). Both orientations were obtained for the 3.7-kb “short” insert (pPR23–1 and pPR23–2), and only one orientation was obtained for the 5-kb insert (pPR27). The plasmid pMJ10 was constructed by inserting a 1.2-kb BamHI fragment from pUC4K, containing a kanamycin resistance gene, into the BamHI site of pPR27.
Mutagenesis vectors were constructed by inserting blunt-ended HindIII (4-kb) fragments, containing the IS1096::kanamycin resistance cassette (Km) derivatives and excised from pYUB285 and pYUB297 (7), into blunt-ended BamHI-cut pPR23 or pPR27 vectors. Five different mutagenesis vectors were obtained, pPR28 to pPR32, according to the delivery vector (pPR23 or pPR27), the transposon (Tn5367 or Tn5368) used, and the orientation of the transposon.
The purC gene was excised from pMJ1 (16) on a 2.5-kb BamHI fragment and inserted into BamHI-cut pACYC184. The resulting vector was named “pMJ100.” The aph cassette from pUC4K conferring kanamycin resistance, present on a 1.2-kb PstI fragment, was cloned into pMJ100 at the single PstI site present in purC. purC::Km was extracted from the resulting pMJ101 vector on a 3.7-kb BamHI fragment, blunt-ended, and ligated into SmaI-cut pXYL4, an E. coli vector containing xylE bracketed by two BamHI sites. The resulting vector was named pMJ102. pMJ103, the construct used for the allelic exchange, was obtained by transferring a 4.7-kb BamHI fragment from pMJ102, containing purC::Km and xylE, into BamHI-cut pPR27.
RESULTS
Design and Testing of a Methodology for the Selection of Insertional Mutants.
Recently, we demonstrated that expression of the sacB gene from B. subtilis is lethal to mycobacteria in the presence of sucrose. sacB can therefore be used as a counterselectable marker (13, 18, 19). We therefore tested whether sacB could be used for the positive selection of insertional mutants. A series of conditionally replicative vectors, combining the counterselective properties of the sacB gene and a mycobacterial thermosensitive origin of replication, was constructed (Fig. 1). These ts-sacB vectors were introduced into M. smegmatis mc2155 by electroporation (17). M. smegmatis transformants, selected at 32°C on 7H10-gentamicin, were grown in 7H9 at 32°C until saturation. The efficiency of the different counterselections was then estimated by plating 100-μl samples of these cultures at different temperatures on 7H10-gentamicin plates with or without 10% sucrose and by counting colony-forming units. Stability of the pAL5000 thermosensitive origin of replication was measured by plating samples at 39°C, the restrictive temperature for replication, without sucrose addition. The efficiency of sacB counterselection was estimated by plating samples at 32°C in the presence of 10% sucrose. By plating on sucrose plates at 39°C, the global counterselection was assessed (Table 1). Each of the counterselective pressures (sucrose and growth temperature) was individually low and led to only a limited loss of the vector. However, when transformants were counterselected for both sacB and the thermosensitive origin of replication, the efficiency of counterselection was extremely high (Table 1). Because gentamicin resistance is not a reliable marker in bacteria of the M. tuberculosis complex, we performed these experiments in M. bovis BCG using pMJ10, a derivative of pPR27 containing a kanamycin resistance gene. Unfortunately, it was not possible to obtain precise data for bacteria of the M. tuberculosis complex because M. bovis BCG (pMJ10) transformants gave microscopic colonies at 39°C, even after 4 weeks of incubation. Nevertheless, as demonstrated (22), thermosensitivity was weaker in mycobacteria of the M. tuberculosis complex than in M. smegmatis. On the other hand, the sucrose selection was more efficient in M. bovis BCG than in M. smegmatis; the measured efficiency of sucrose counterselection was 5.5 × 10−5. Therefore, it was likely that the efficiency of the dual counterselection in M. tuberculosis should be at least as high as the sucrose counterselection alone. We observed that the dual counterselection in M. bovis BCG (pMJ10) led to an almost complete clearance of the background obtained when the sucrose selection was applied alone (data not shown). This suggested that ts-sacB vectors could be used to deliver a transposon or a mutated allele into the chromosome of M. tuberculosis, allowing the construction of insertional mutant libraries or gene exchange mutants, respectively. This protocol of selection was used for all subsequent mutagenesis experiments. Because transformants are grown under permissive conditions, problems caused by low transformation efficiencies are avoided. During this step of replication, mutants arising by allelic exchange or transposition can accumulate, overcoming problems caused by low frequencies of allelic exchange and transposition. Finally, the great majority of the clones that still contain the vector are eliminated, strongly increasing the proportion of mutants among the survivors.
Figure 1.
Design of ts-sacB vectors for positive selection of rare genetic events (pPR27 is shown as an example). Only single restriction sites that can be used for the subsequent cloning of a transposon or a mutant allele are shown.
Table 1.
Effect of sucrose and temperature on growth of M. smegmatis transformed with pPR27
| Growth conditions | cfu/100 μl | Counterselection efficiency |
|---|---|---|
| 32°C | 1.2⋅107 | |
| 39°C | 5000 | 4.2· 10−4 |
| sucrose at 32°C | 3000 | 2.5· 10−4 |
| sucrose at 39°C | 6 | 5· 10−7 |
Identical results were obtained for pPR23 (data not shown).
Construction of Transposition Mutant Libraries of M. tuberculosis and M. bovis BCG.
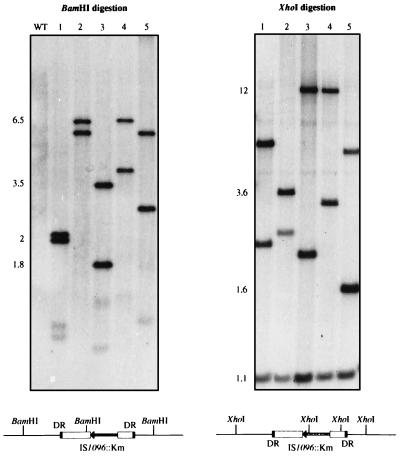
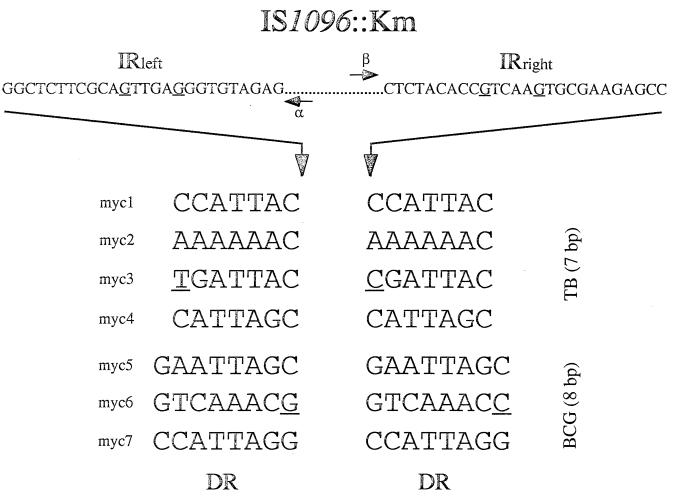
ts-sacB vectors were first tested for transposon mutagenesis in bacteria of the M. tuberculosis complex. The devised selection protocol implies that the delivery vector, containing the counterselectable sacB gene, should be lost upon transposition such that transposition mutants can be positively selected. In other words, the transposon to be used should transpose in a conservative fashion (23). The only known mycobacterial mobile element suitable for our system was IS1096 (15); it is not found in the chromosomes of the M. tuberculosis complex mycobacteria, and its transposition is random and mostly conservative. Moreover, derivatives of IS1096, with a Km inserted into the mobile element (IS1096::Km), are able to transpose in M. bovis BCG (7). Two different IS1096 derivatives, Tn5367 and Tn5368, were cloned into pPR23 and pPR27 delivery vectors, giving a series of mutagenesis vectors that were introduced in M. tuberculosis 103 and M. bovis BCG Pasteur by electroporation. The selection strategy described above was used. Putative insertional mutants, in which IS1096::Km may have transposed onto the chromosome, were selected at 39°C on 7H10-kanamycin + 2% sucrose plates. When an initial inoculum of 107 bacteria was plated, 104 and 105 colonies were obtained for M. tuberculosis and the vaccinal strain, respectively. Five randomly picked M. tuberculosis (pPR32) transformants were analyzed by Southern blot, using pPR32 as a probe. Two hybridizing fragments of varying sizes were expected when BamHI was used (Fig. 2). XhoI cuts twice in IS1096::Km, so transposition mutants were expected to present three hybridizing fragments (Fig. 2): one fragment of conserved length, internal to the mobile element, and two fragments the sizes of which would differ between the mutants provided that the transposition was random. The hybridization patterns for all of the clones tested were in agreement with the transposition of Tn5368 onto the chromosome of the tubercle bacillus (Fig. 2). This was also confirmed with other restriction enzymes (data not shown). The same blot was analyzed with the delivery vector as a probe, i.e., pPR27, which contains no inserted transposon. No hybridization signal was detected (data not shown), confirming that the delivery vector had been lost during transposition. Independent transposition experiments were repeated at least five times in both M. tuberculosis and M. bovis BCG, and similar Southern hybridization results were obtained for all of the mutagenesis vectors. More than 100 mutants were examined by Southern blot analysis, and >95% of the clones resulted from the transposition of IS1096::Km onto the chromosome. However, it should be noted that, in some experiments, several clones exhibited the same hybridization patterns, suggesting that they were siblings. This was not unexpected because the mutagenesis protocol contains a step during which possible mutants are able to replicate. Nevertheless, most of the hybridization patterns of different clones were unique, suggesting that IS1096 transposition occurred at random as described (7). To confirm the randomness of the transposition, several insertion sites were cloned and sequenced. Direct repeats (DR) bracketing the transposon were found, confirming that the clones indeed resulted from transposition events (Fig. 3). Two clones, myc3 and myc6, presented imperfect DRs probably resulting from replication errors. Moreover, DRs were of different lengths between M. tuberculosis and M. bovis BCG mutants: 7 bp and 8 bp, respectively. This is, to our knowledge, the first report of such a phenomenon: DR length being specific for the strain in which transposition occurs. The mechanism responsible for this strain specificity is so far unknown. Important to note, despite a preference for A+T rich targets, IS1096 does not appear to display site-specificity; all of the analyzed insertion sites were different (Fig. 3). These results confirmed that the ts-sacB vectors were indeed suitable for delivering transposons onto the chromosome of bacteria of the M. tuberculosis complex. Moreover, the methodology was reliable and fully reproducible, enabling the construction of insertional mutant libraries containing >106 different transposition mutants.
Figure 2.
Southern blot analysis of representative M. tuberculosis::Tn5368 clones and expected schematic hybridization patterns for a transposition mutant. Five mutants were picked at random (clones 1–5). M. tuberculosis 103 DNA (WT) was included as a control and as expected showed no hybridization signal. Genomic DNA was digested with BamHI or XhoI and probed for hybridization with pPR32, a vector consisting of the Tn5368 transposon cloned into the BamHI site of pPR23. Molecular masses are indicated in kilobases.
Figure 3.
Sequences of cloned insertion sites for several M. tuberculosis and M. bovis BCG transposition mutants. Approximately 500 bp of DNA flanking the transposon was sequenced using IS1096 outward primers α and β, but only DR are shown. Imperfectly repeated nucleotides are underlined.
Gene Exchange Mutagenesis of the purC Gene of M. tuberculosis.
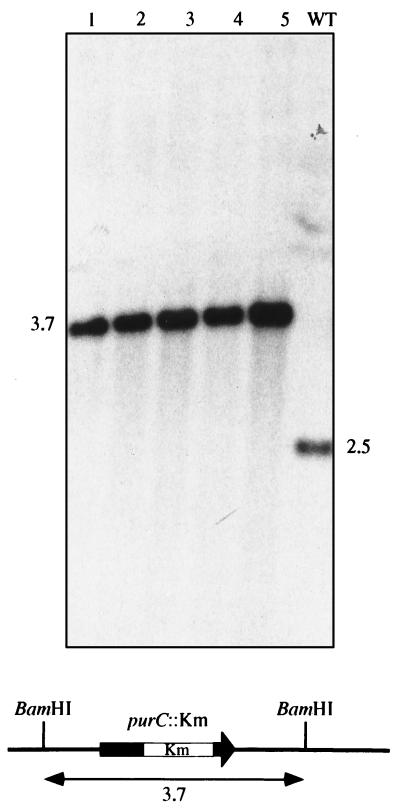
The selection protocol used for M. tuberculosis transposon mutagenesis also seemed attractive for the positive selection of gene exchange mutants. Currently, none of the described strategies appears suitable for mutagenesis of every gene in M. tuberculosis complex strains (11–13). The systems available are very dependent on the target gene and have proved far less efficient for several “refractory” genes such as purC, which we were unable to mutagenize in M. tuberculosis (unpublished data). Because M. tuberculosis purine auxotrophic mutants may have a vaccinal potential (24), the purC gene from the purine biosynthetic pathway was a perfect candidate for testing our new tool (16). A mutated allele, purC::Km, was inserted into pPR27 along with xylE as a reporter gene. The gentamicin resistance gene is not a reliable marker in M. tuberculosis, so we reasoned that this reporter activity could facilitate the screening by discriminating possible allelic exchange mutants that have lost the xylE gene, from sacB mutants with the whole vector integrated into the chromosome and that are thus phenotypically XylE+. Indeed, xylE expression in mycobacteria can easily be tested by spraying colonies on plates with a solution of catechol and by observing a bright yellow coloration (25). Plasmid pMJ103 was introduced in M. tuberculosis by electroporation, and transformants were selected at 32°C on 7H10-kanamycin. Several transformants were grown in liquid culture supplemented with hypoxanthin, a purine precursor. The culture was then plated at 39°C on 7H10-kanamycin + 2% sucrose + hypoxanthin plates. With an initial inoculum of 107 colonies, 200 transformants were obtained on counterselective plates. All presented the expected phenotype for allelic exchange mutants: Sucr, Kmr, XylE−. The phenotypic analysis confirmed that they were purine auxotrophs because they were not able to grow on Sauton medium, a synthetic medium containing no purine bases, without the addition of hypoxanthin. To unambiguously confirm that the selected clones were allelic exchange mutants, several colonies were grown in 7H9 supplemented with hypoxanthin. Genomic DNA was extracted and analyzed by Southern blotting using the purC gene as a probe (Fig. 4). M. tuberculosis 103 DNA, which was included as a control, showed one hybridizing fragment of 2.5 kb. As expected for allelic exchange mutants, all of the clones presented a single hybridizing fragment ≈1.2 kb longer than that in the wild-type strain (Fig. 4). This 1.2-kb increase corresponded to the size of the Km that was inserted into the mutated allele. Therefore, all of the tested transformants, selected on counterselective plates, were indeed allelic exchange mutants. This confirmed that ts-sacB delivery vectors, in addition to being useful for transposon mutagenesis, are also highly efficient for gene exchange mutagenesis.
Figure 4.
Southern blot analysis of M. tuberculosis purC mutants and expected schematic pattern of hybridization for an allelic exchange mutant. Five auxotrophic mutants were picked at random (clones 1–5). Chromosomal DNA was digested with BamHI and probed for hybridization with the pMJ103 vector. M. tuberculosis 103 DNA was included as a control (WT). Molecular masses are indicated in kilobases.
DISCUSSION
A single, very simple system that can be used for easy mutagenesis of M. tuberculosis either by allelic exchange or by transposition was designed. ts-sacB vectors were successfully used to deliver a mycobacterial transposon into the chromosome of two bacteria of the M. tuberculosis complex: M. tuberculosis and M. bovis BCG. The great majority of the selected clones (>95%) were insertional mutants, with a copy of IS1096::Km inserted in their chromosome. Moreover, Southern blot analysis (Fig. 2) and sequencing of the flanking regions (Fig. 3) indicated that the mutant libraries, numbering >106 mutants and containing transposons inserted in various sites, were representative. It seems likely that the pool of mutants contains clones in which each of the nonessential mycobacterial genes, numbering 3000, have been mutated at least once (8). These libraries will therefore be extremely useful for the identification of avirulent mutants, possibly unable to invade epithelial cells or to replicate in macrophages, in which essential virulence factors have been disrupted (unpublished work). In addition, using purC, a gene that we were previously unable to mutate in M. tuberculosis, we demonstrated that ts-sacB vectors can also be used for allelic exchange mutagenesis. All of the clones obtained after a double selection were indeed allelic exchange mutants. The M. tuberculosis purine auxotrophic mutant, which might display attenuated virulence (currently being investigated) may present candidate vaccinal potential (24) and may also be used for the development of an in vivo expression technology allowing the selection of mycobacterial genes preferentially expressed in vivo (26). Provided that its function is dispensable to the cell, it is reasonable to assume that the same protocol should allow mutagenesis of virtually every gene of M. tuberculosis through gene exchange.
The tool we have developed should greatly contribute to the genetic analysis of M. tuberculosis pathogenicity following Koch’s molecular postulates (2, 3); it allows creation of defined mycobacterial mutants either by allelic exchange or transposon mutagenesis, both of which were previously difficult or even unfeasible. It opens the way not only to studying the roles in pathogenicity of defined mycobacterial genes that may or not present similarities to known virulence factors from other bacterial pathogens but also to the rational construction of attenuated strains that could be more effective than the BCG as antituberculous vaccines.
Acknowledgments
We are grateful to A. Edelman and T. Msadek for critical reading of the manuscript. We thank J. Rauzier for automated sequencing. V.P. and M.J. are recipients of Pasteur–Weizmann and Fondation Mérieux fellowships, respectively. This work was supported by a European Economic Community Biotech program grant (BIO-CT92-0520), the National Institutes of Health Grant AI 35207, and Institut Pasteur.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TB, tuberculosis; DR, direct repeat; Km, kanamycin resistance cassette; BCG, Bacille Calmette–Guérin.
References
- 1.Bloom B R, Murray C J L. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Falkow S. Rev Infect Dis. 1988;10:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs W R., Jr Immunobiology. 1992;184:147–156. doi: 10.1016/S0171-2985(11)80472-9. [DOI] [PubMed] [Google Scholar]
- 4.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley L W. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 5.Pascopella L, Collins F M, Martin J M, Lee M H, Hatfull G F, Stover C K, Bloom B R, Jacobs W R., Jr Infect Immunol. 1994;62:1313–1319. doi: 10.1128/iai.62.4.1313-1319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins D M, Kawakami R P, de Lisle G W, Pascopella L, Bloom B R, Jacobs W R., Jr Proc Nat Acad Sci USA. 1995;92:8036–8040. doi: 10.1073/pnas.92.17.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAdam R, Weisbrod T R, Martin J, Scuderi J D, Brown A M, Cirillo J D, Bloom B R, Jacobs W R., Jr Infect Immunol. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs W R, Jr, Bloom B R. In: Tuberculosis. Bloom B R, editor. Washington, D.C.: Am. Soc. Microbiol.; 1994. pp. 253–268. [Google Scholar]
- 9.Reyrat J-M, Berthet F-X, Gicquel B. Proc Natl Acad Sci USA. 1995;92:8768–8772. doi: 10.1073/pnas.92.19.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azad A K, Sirakova T D, Rogers L M, Kolattukudy P E. Proc Natl Acad Sci USA. 1996;93:4787–4792. doi: 10.1073/pnas.93.10.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norman E, Dellagostin O A, McFadden J, Dale J W. Mol Microbiol. 1995;16:755–760. doi: 10.1111/j.1365-2958.1995.tb02436.x. [DOI] [PubMed] [Google Scholar]
- 12.Balasubramanian V, Pavelka Jr M S, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelicic V, Reyrat J-M, Gicquel B. FEMS Microbiol Lett. 1996;144:161–166. doi: 10.1111/j.1574-6968.1996.tb08524.x. [DOI] [PubMed] [Google Scholar]
- 14.Guilhot C, Otal I, van Rompaey I, Martín C, Gicquel B. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cirillo J D, Barletta R G, Bloom B R, Jacobs W R., Jr J Bacteriol. 1991;173:7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson M, Berthet F-X, Otal I, Rauzier J, Martin C, Gicquel B, Guilhot C. Microbiology. 1996;142:2439–2447. doi: 10.1099/00221287-142-9-2439. [DOI] [PubMed] [Google Scholar]
- 17.Snapper S B, Melton R E, Mustapha S, Kieser T, Jacobs W R., Jr Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 18.Pelicic V, Reyrat J-M, Gicquel B. J Bacteriol. 1996;178:1197–1199. doi: 10.1128/jb.178.4.1197-1199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelicic V, Reyrat J-M, Gicquel B. Mol Microbiol. 1996;20:919–925. doi: 10.1111/j.1365-2958.1996.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 20.Guilhot C, Gicquel B, Martín C. FEMS Microbiol Lett. 1992;98:181–186. doi: 10.1016/0378-1097(92)90152-e. [DOI] [PubMed] [Google Scholar]
- 21.Quandt J, Hynes M F. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 22.Guilhot, C. (1995) Ph.D. dissertation, Paris 7 University (Paris).
- 23.McAdam R, Guilhot C, Gicquel B. In: Tuberculosis. Bloom B R, editor. Washington, D.C.: Am. Soc. Microbiol.; 1994. pp. 199–216. [Google Scholar]
- 24.Fields P I, Swanson R V, Haidaris C G, Heffron F. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curcic R, Dhandayuthapani S, Deretic V. Mol Microbiol. 1994;13:1057–1064. doi: 10.1111/j.1365-2958.1994.tb00496.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahan M J, Slauch J M, Mekalanos J J. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]