Abstract
Transposon mutagenesis provides a direct selection for mutants and is an extremely powerful technique to analyze genetic functions in a variety of prokaryotes. Transposon mutagenesis of Mycobacterium tuberculosis has been limited in part because of the inefficiency of the delivery systems. This report describes the development of conditionally replicating shuttle phasmids from the mycobacteriophages D29 and TM4 that enable efficient delivery of transposons into both fast- and slow-growing mycobacteria. These shuttle phasmids consist of an Escherichia coli cosmid vector containing either a mini-Tn10(kan) or Tn5367 inserted into a nonessential region of the phage genome. Thermosensitive mutations were created in the mycobacteriophage genome that allow replication at 30°C but not at 37°C (TM4) or 38.5°C (D29). Infection of mycobacteria at the nonpermissive temperature results in highly efficient transposon delivery to the entire population of mycobacterial cells. Transposition of mini-Tn10(kan) occurred in a site-specific fashion in M. smegmatis whereas Tn5367 transposed apparently randomly in M. phlei, Bacille Calmette–Guérin (BCG), and M. tuberculosis. Sequence analysis of the M. tuberculosis and BCG chromosomal regions adjacent to Tn5367 insertions, in combination with M. tuberculosis genomic sequence and physical map data, indicates that the transpositions have occurred randomly in diverse genes in every quadrant of the genome. Using this system, it has been readily possible to generate libraries containing thousands of independent mutants of M. phlei, BCG, and M. tuberculosis.
In April 1993, tuberculosis was declared a global health emergency—the first such designation in the history of the World Health Organization. The distinction is regrettably justified because tuberculosis remains one of the largest burdens of disease and death in the world (1, 2) due in part to the increased susceptibility of HIV-infected individuals and the ominous emergence of multidrug resistant strains in both industrialized and developing countries. It is our view that effective new tuberculosis control and prevention strategies will require additional knowledge of the causative agent and its interaction with the human host.
To systematically delineate virulence determinants, identify the metabolic pathways, and discover novel drug targets for Mycobacterium tuberculosis, a methodology generating libraries of mutants will be essential. Although mutant isolation and gene transfer strategies have been successfully used for fast-growing nonvirulent mycobacteria, such as M. smegmatis (3, 4), determining the genetic basis of phenotypes for M. tuberculosis has been frustrated by the lack of a natural gene transfer system in this pathogen. Furthermore, traditional mutational analyses based on the characterization of colonies arising from single cells after treatment with DNA-damaging agents is of limited value for slow-growing mycobacteria because the frequency of mutants is very low, multiple mutations occur in the same cells, and the mycobacteria tend to clump.
Transposon mutagenesis has been successfully used in diverse genera of bacteria (5, 6). The first transposition events in M. smegmatis were reported using Tn610 (7), followed by transposons engineered from insertion sequences (IS)900 and IS986 (8, 9). Transposition in Bacille Calmette-Guérin (BCG) (12) was reported using a transposon constructed from the insertion element IS1096 (10). Remarkably, the only reports of successful isolation of auxotrophic mutants for mycobacteria of the M. tuberculosis group used insertional mutagenesis systems: illegitimate recombination (11), transposon mutagenesis (12), and allelic exchange (13). A very promising approach to deliver transposons into M. smegmatis is the use of a conditionally replicating vector that is able to replicate at 30°C but not at 37°C (14). A library of 30,000 Tn611 insertion mutants was obtained from three independent experiments yielding 80 auxotrophic mutants with 15 different phenotypes. However, this system had not yet been applied to the slow-growing mycobacteria such as M. tuberculosis.
Conditionally replicating phage systems have proven to be very efficient systems for transposon mutagenesis in numerous bacterial species (5). One of the great advantages of a phage delivery system is that essentially every cell in the bacterial population can be infected with the transposon-carrying phage, generating large numbers of independent mutants. Shuttle phasmid vectors, chimeric molecules that replicate in Escherichia coli as plasmids and in mycobacteria as phages, were the first recombinant DNA vectors engineered for mycobacteria (15). Various phasmids, constructed from different mycobacteriophages, such as TM4, L1, and D29, have proven useful for the development of transformation systems for mycobacteria (16) and for the development of luciferase reporter phages for rapid diagnosis and drug susceptibility testing of M. tuberculosis clinical isolates (17, 18).
In this report, we describe the isolation of conditionally replicating D29 and TM4 shuttle phasmid vectors that can replicate in M. smegmatis at 30°C but fail to replicate and lyse the host cells at 38.5°C (D29-based phasmids) or 37°C (TM4-based phasmids). With these phasmids engineered to deliver the transposons Tn5367 or mini-Tn10(kan), we were able to generate efficiently transposon libraries of M. phlei, BCG, and M. tuberculosis mutants.
MATERIALS AND METHODS
Bacterial Strains, Media, and Culture Methods.
The mycobacterial strains used in this study are listed in Table 1. The E. coli strains, DH5α or HB101, used for cloning hosts were grown as described (19). For preparation of electrocompetent cells for transfections, M. smegmatis mc2155 or mc21255 were grown in Luria–Bertani plus Tween 80 0.05% broth (20). Mycobacteriophages were propagated in mc2155 as described (21). When required, the following antibiotics were used at the specified concentrations: carbenicillin (50 μg/ml) and kanamycin (25 μg/ml for E. coli; 20 μg/ml for mycobacteria); BCG, M. tuberculosis Erdman strain, and other mycobacteria were grown in Middlebrook 7H9 broth enriched with albumin (fraction V) 0.5%, dextrose 0.2%, sodium chloride 0.85%, and Tween 80 0.05% (M-ADC-TW) broth (21). For transposon delivery experiments, BCG and M. tuberculosis were grown in M-ADC-TW broth without glycerol.
Table 1.
Mycobacterial strains used in this study
| Species | Strain | Genotype or description | Reference or source |
|---|---|---|---|
| M. aurum | mc218 | W. Jones, CDC | |
| M. chelonae | mc221 | W. Jones, CDC | |
| M. phlei | mc219 | Phage host | W. Jones, CDC |
| M. phlei | ATCC354 | NTCC54 | ATCC |
| M. phlei | ATCC11758 | TMC1548, type strain | ATCC |
| M. phlei | ATCC27086 | Jahasz F89 | ATCC |
| M. phlei | ATCC27206 | SN109 | ATCC |
| M. fortuitum | ATCC6841 | TMC1529 | ATCC |
| M. fortuitum | ATCC35755 | TMC1545 | ATCC |
| M. flavescence | ATCC14474 | TMC1541 | ATCC |
| M. smegmatis | mc2155 | ept-1 | ref. 23 |
| M. smegmatis | mc21255 | ept-1 rpsL4 | ref. 20 |
| M. smegmatis | ATCC359 | Alias M. butiricum | ATCC |
| M. smegmatis | ATCC11759 | Penso strain Milch | ATCC |
| M. smegmatis | ATCC23032 | Runyon Special 9 | ATCC |
| M. smegmatis | ATCC27204 | SN2 | ATCC |
| M. smegmatis | ATCC27205 | SN38 | ATCC |
| M. smegmatis | ATCC35797 | TMC1515 | ATCC |
| M. smegmatis | ATCC37798 | TMC1533 “R. Koch” | ATCC |
| M. vaccae | ATCC15483 | TMC1526 SN920 | ATCC |
| M. vaccae | ATCC23014 | SN923 | ATCC |
| M. tuberculosis | Erdman | Virulent isolate | F. Collins, FDA |
| BCG | Pasteur strain | Staten Serum Institut |
CDC, Centers for Disease Control; ATCC, American Type Culture Collection; FDA, Food and Drug Administration.
Isolation of Temperature-Sensitive Mycobacteriophages.
D29 shuttle phasmids were propagated in M. smegmatis mc2155. Samples of phages (109 pfu/ml) were mutagenized with hydroxylamine as described (22) using conditions that yielded 0.1% viable surviving phages. These were plated on M. smegmatis mc2155 and incubated for 24 h at 30°C until very small plaques appeared. Plates were then shifted at 42°C and incubated for another 24–36 h. Phage plaques that remain very small after this incubation period were screened for their abilities to form plaques at 30, 37, and 42°C. Phage clones forming plaques at 30°C, but not at 42°C, were clone-purified, amplified to a high titer, and analyzed for reversion frequency. Shuttle phasmids from either D29 or PH101 were prepared as described (15, 17, 18).
Recombinant DNA Methodologies.
Cosmids, phasmids, transposons, and phages used in this study are listed in Table 2. DNA manipulations were done essentially as described (19). High molecular weight chromosomal DNA from BCG or M. tuberculosis for generating cosmid libraries was purified as described (13).
Table 2.
Phages, cosmids, shuttle plasmids, and transposons used in this study
| Description | Reference | |
|---|---|---|
| Transposons | ||
| Tn5367 | IS1096-derived transposon containing kanr gene | 12 |
| Mini-Tn10(kan) | Derivative of Tn10-containing aph gene | 5 |
| Cosmids | ||
| pYUB328 | ColE1 ampr exciseable double cos vector | 13 |
| pYUB552 | Minimal (2.4-kb) pYUB328 cosmid derivative | This work |
| pYUB553 | pYUB552::Tn5367 | This work |
| pYUB554 | pYUB552::mini-Tn10(kan) | This work |
| Phages | ||
| λ | cI857, Sam10 | |
| D29 | Wild type | 15 |
| TM4 | Wild type | 15 |
| PH101 | ts mutant of TM4 that fails to form plaques at 42°C | This work |
| Shuttle phasmids | ||
| phAE60 | D29 Δ (45440-48108 bp)::pYUB328 | This work |
| phAE65 | ts mutant of phAE60 that fails to replicate at 42°C | This work |
| phAE70 | ts mutant of phAE65 that fails to replicate at 38.5°C | This work |
| phAE71 | ts mutant of phAE65 that fails to replicate at 38.5°C | This work |
| phAE72 | ts mutant of phAE65 that fails to replicate at 38.5°C | This work |
| phAE73 | ts mutant of phAE65 that fails to replicate at 38.5°C | This work |
| phAE76 | ts mutant of phAE65 that fails to replicate at 38.5°C | This work |
| phAE87 | PH101::pYUB328 that fails to replicate at 37°C | This work |
| Conditionally replicating transposon delivery shuttle plasmids | ||
| phAE77 | phAE70::pYUB553 | This work |
| phAE78 | phAE70::pYUB554 | This work |
| phAE94 | phAE87::pYUB553 | This work |
Transposon Mutagenesis.
Fast-growing mycobacteria were cultured in LBT to an OD at A600 of 1.0 (≈2 × 108 cfu/ml). BCG and M. tuberculosis were grown for 7–10 days after inoculation of 10 ml of the starter culture into a 100-ml roller bottle culture (A600 = 1.0) in M-ADC-TW without glycerol plus 0.4% casamino acids. Ten milliliters of cultures was concentrated by centrifugation and resuspended in 1 ml of MP buffer (50 mM Tris·HCl, ph 7.6/150 mM NaCl/2mM CaCl2) (21). The cells were prewarmed at the nonpermissive temperature and then were mixed with 2 × 1010 pfu/ml [multiplicity of infection (MOI) = 10]. The cell–phage mixture was incubated at the nonpermissive temperature for 30 min for the fast-growing mycobacteria or 4–6 hours for the slow-growing mycobacteria. After completion of the adsorption time, 2 ml of prewarmed “adsorption-stop buffer” (MP buffer containing 20 mM sodium citrate and 0.2% Tween 80) was added to prevent further phage infections. Samples of this mixture were plated on prewarmed tryptic soy agar plates for the fast-growing mycobacteria or M-ADC-TW-based agar media containing 0.1% Tween 80, 0.4% casamino acids, 40 μg/ml l-tryptophan, and 20 μg/ml kanamycin for the slow-growing mycobacteria. Transposition frequency was estimated as a number of kanamycin-resistant (kanr) colonies per total number of input cells.
Southern Blotting and DNA Sequencing.
Southern blotting was done by the alkali-denaturing procedure. DNA was transferred to Biotrans nylon membranes (ICN) by the capillary method. Hybridization and detection were done as recommended by the manufacturer (enhanced chemiluminescence, Amersham). Sequence analysis was performed using the Applied Biosystems Prism Dye Terminator Cycle Sequencing Core kit with AmpliTaq DNA polymerase (Perkin–Elmer) and an Applied Biosystems 377 automated DNA sequencer. Chromosomal DNA sequence at the junction of the transposon insertions was obtained by PCR sequencing outwards from the transposon using primers HOPS1 (5′-GGCGTAGGAACCTCCATCATC-3′) and HOPS2 (5′-CTTGCTCTTCCGCTTCTTCTCC-3′) with cosmid DNA as a template. The sequence thus obtained (400–600 bp) was used to blast-search the M. tuberculosis Sanger Sequence database or GenBank database.
RESULTS
Development of Conditionally Replicating Shuttle Phasmids.
Our goal was to develop conditionally replicating mycobacteriophage vectors for the delivery of transposons to a broad range of mycobacteria, including the slow-growing M. tuberculosis and BCG (Fig. 1). We chose to construct a phasmid vectors with the exciseable cosmid pYUB328 (13) because such vectors provide an easy means to introduce transposons into nonessential regions of a mycobacteriophage. Shuttle phasmids from D29 were first constructed, and one such phasmid, phAE60, which had a maximal deletion within the D29 phage, was subjected to hydroxylamine mutagenesis. From the initial screen of 2000 mutants only phages unable to replicate at 42°C were observed. One such mutant, phAE65, was subjected to further mutagenesis, and a set of independent mutants was identified that failed to form plaques at 38.5°C (Table 2). The mutations conferring thermosensitive growth reverted at frequencies of 10−4 at 38.5°C and <10−7 at 42°C. Six mutants were tested for their inability to kill M. smegmatis when mixed at MOI of 10 and incubated at the nonpermissive temperature. One such mutant, phAE70, showing the lowest reversion frequency and inability to kill M. smegmatis when incubated at the nonpermissive temperature (38.5°C), was chosen as the D29-based delivery phage.
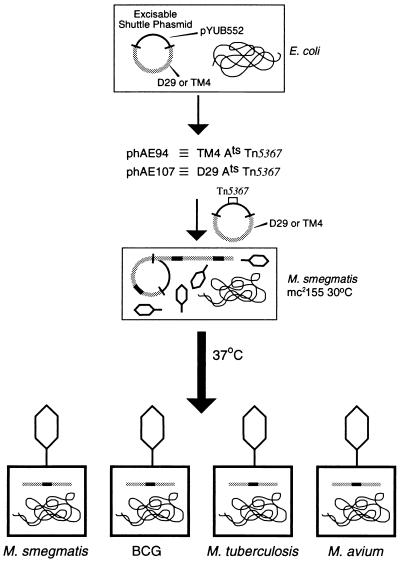
Figure 1.
Transposon delivery in mycobacteria using conditionally replicating shuttle phasmids. Shuttle phasmids are mycobacteriophage molecules into which an E. coli cosmid has been inserted in a nonessential region. These molecules can thus replicate in E. coli as cosmids and replicate in mycobacteria as phages. The cosmid is flanked by restriction sites that are not found in the shuttle phasmid, thus allowing for simple excis-ion of the cosmid. A cosmid containing a transposon is readily cloned into the phage backbone by cosmid cloning in E. coli. Cosmids are isolated from E. coli and transfected into M. smegmatis cells by electroporation. At permissive temperatures (30°C), shuttle phasmids undergo growth as a lytic mycobacteriophage to high titers. Infection of various mycobacteria with the mycobacteriophages containing the transposons results in the delivery to every cell in a population of mycobacterial cells. The temperature-sensitive mutations in the mycobacteriophages prevent phage propagation, which allows for transpositions into the mycobacterial chromosome to occur. The broad host ranges of D29 and TM4 mycobacteriophages allow for delivery of transposons to a variety of different mycobacterial species, including BCG and M. tuberculosis.
For the construction of the TM4 shuttle phasmid transposon delivery vector, we first mutagenized the wild-type TM4 phage and isolated the temperature-sensitive mutant PH101. This mutant failed to form plaques at 37°C, had a reversion frequency <10−7, and failed to kill M. smegmatis at 37°C at an MOI of 10. As in the case of D29-based phasmids, two independent rounds of mutagenesis were needed. A shuttle phasmid was engineered from this phage and was designated phAE87. Both phAE70 and phAE87 were used for the construction of transposon delivery vectors.
Delivery of Transposons in Fast-Growing Mycobacteria Using Conditionally Replicating Shuttle Phasmids.
Transposon constructs could be readily introduced into either phAE70 or phAE87 by replacing the pYUB328 cosmid with a cosmid containing the transposon of interest. Initial attempts to introduce transposon constructs, engineered in pYUB328, into the D29 shuttle phasmids were unsuccessful, most likely because of the packaging constraints of this phage. To further maximize the cloning capacities, a cosmid vector, pYUB552, was engineered to contain 2.1 kb less DNA than pYUB328. Tn5367 (12) or mini-Tn10(kan) (5) was cloned into this cosmid to generate pYUB553 and pYUB554, respectively. Replacement of the pYUB328 in phAE70 with pYUB554 generated phAE78. Infection of M. smegmatis mc2155 cells with phAE78 at MOI of 10 yielded kanr colonies at frequencies slightly >10−6 in four independent experiments (Table 3). Southern blot analysis revealed that chromosomal DNA from the kanr colonies contained no D29 phage sequences (data not shown) but did contain the kanr cassette of the mini-Tn10(kan) (Fig. 2A). Unfortunately, mini-Tn10(kan) appeared to integrate in a site-specific fashion in M. smegmatis and, thus, would not be useful for generating libraries of random insertional mutants. Nevertheless, these results confirmed that the D29 conditionally replicating shuttle phasmids could be used efficiently to deliver transposons to mycobacterial cells.
Table 3.
Transposition of mini-Tn10(kan) and Tn5367 in fast-growing mycobacteria
| Species | Strain | Copies of IS1096 | Transpositions | |
|---|---|---|---|---|
| Mini-Tn10(kan) | ||||
| via: phAE78 | ||||
| M. smegmatis | mc2155 | 11 | 3600 | |
| Tn5367 via: | ||||
| phAE77 | phAE94 | |||
| M. smegmatis | mc2155 | 11 | >10 | >10 |
| M. smegmatis | ATCC359 | 3 | 80 | 215 |
| M. smegmatis | ATCC11759 | 6 | 0 | 0 |
| M. smegmatis | ATCC23032 | 9 | 330 | 345 |
| M. smegmatis | ATCC27204 | 5 | 0 | 0 |
| M. smegmatis | ATCC27205 | 14 | 0 | 0 |
| M. smegmatis | ATCC35797 | 11 | 85 | 260 |
| M. smegmatis | ATCC35798 | 9 | 285 | 425 |
| M. aurum | mc218 | None | 0 | 0 |
| M. chelonae | mc221 | None | 0 | 0 |
| M. phlei | mc219 | None | 2 | 2000 |
| M. phlei | ATCC354 | None | 0 | 3 |
| M. phlei | ATCC11758 | None | 0 | 0 |
| M. phlei | ATCC27086 | None | 0 | 0 |
| M. phlei | ATCC27206 | None | 0 | 1 |
| M. fortuitum | ATCC6841 | None | 0 | 0 |
| M. vaccae | ATCC15483 | None | 0 | 0 |
| M. vaccae | ATCC23014 | None | 0 | 0 |
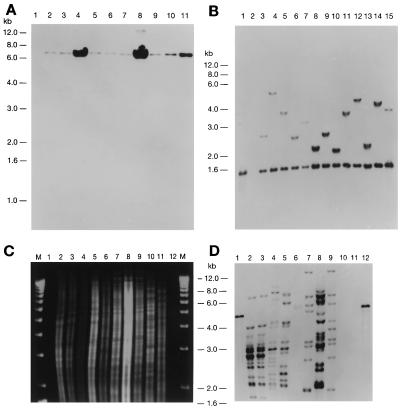
Figure 2.
Southern blot analysis of transposon mutants. (A) kanr M. smegmatis mc2 155 mutagenized with mini-Tn10(kan) using phAE78 as a delivery phage. Chromosomal DNAs were isolated from 11 independent mutants, digested with PstI (which does not cleave in the transposon), and hybridized with a DNA fragment containing the gene that confers kanr. (B) kanr M. phlei cells mutagenized with Tn5367 using phAE77 as a delivery phage. Chromosomal DNA digested with PstI and ApaLI and hybridized with the aph gene. Both restriction sites are located within the transposon, so a common band is generated as well as the random fragment generated from the insertion of Tn5367 into the M. phlei chromosome. Lane 1 contains pYUB553 digested with PstI and ApaLI. (C and D) Southern blot analysis of IS1096 in fast-growing mycobacteria. Ethidium bromide-stained agarose gel of chromosomal DNAs of various M. smegmatis strains (lanes 2–10); M. phlei (lane 11) and M. vaccae (lane 12) were digested with ApaLI (C) and hybridized with tnpA of IS1096 (D).
Tn5367 represented an attractive transposon for mutagenesis of mycobacteria because we had shown previously that it transposed in BCG in a relatively random fashion by a nonreplicative mechanism (12). pYUB553 was used to replace pYUB328 into both the D29 and TM4 conditionally replicating shuttle phasmids to generate phAE77 and phAE94, respectively. These vectors were used to deliver Tn5367 into mc2 155 cells at 38.5°C (phAE77) or 37°C (phAE94). Very few kanr colonies were obtained. Southern analysis suggested that transposition had occurred because hybridization of the kanr colonies with the aph gene yielded fragments of various sizes (data not shown). We hypothesized that the low frequency of transposition of Tn5367 in mc2 155 might be a result of a negative regulation resulting from the 11 copies of IS1096. Searching for an alternative fast-growing mycobacterial host, we screened various strains of M. smegmatis, M. aurum, M. fortuitum, M. chelonei, M. phlei, and M. vaccae. Transposition in three different M. smegmatis strains and one M. phlei [mc2 19] strain were observed whereas no or very low frequency of transposition occurred in the others (Table 3). In subsequent experiments infecting mc2 19 cells with phAE94 at a MOI of 1, we routinely obtained >2000 kanr clones/108 input cells. Southern analysis revealed that the transposition occurred in a random fashion in M. phlei (Fig. 2B) and in the three M. smegmatis strains (data not shown). The inability to observe transpositions in some strains such as M. fortuitum or M. chelonei is likely a result of the inability of the phages to infect these strains. The ability of phAE77 and phAE94 to form plaques on the fast-growing strains at permissive temperature was shared by all of the strains that yielded transpositions but was not sufficient to predict the transposition efficiency. Nor was the number of copies of IS1096 predictive of transposition efficiency because an M. smegmatis strain with 11 copies yielded as many transpositions as one strain with two and a M. phlei strain that had none (Fig. 2 C and D). Of interest, the presence of IS1096 appears to be uniquely diagnostic for M. smegmatis strains; all eight strains that had the insertion sequence (Fig. 2 C and D) yielded a common restriction fragment length polymorphism after PCR amplification of the hsp60 gene product (ref. 24 and data not shown).
Delivery of Transposons in Slow-Growing Mycobacteria Using Conditionally Replicating Shuttle Phasmids.
For slow-growing mycobacteria, the expression time required for optimal transposition after phage infection was unknown. Therefore, we tested the effect of time of incubation with the phage before plating on kanamycin-containing media. The maximal number of kanr colonies was obtained after 4 h of incubation of BCG with phAE77 or M. tuberculosis using phAE94 (Table 4). Surprisingly, phAE94 yielded no or relatively few transposon mutants in BCG. Using M. tuberculosis, in three independent experiments, phAE94 yielded 1700, 3500, and 8000 transposon mutants, respectively. In contrast, phAE77 yielded relatively few transposon mutants in M. tuberculosis whereas in BCG the number of the kanr colonies was consistently higher than 104. Southern analyses of chromosomal DNA from 20 randomly picked kanr colonies of either BCG or M. tuberculosis revealed a random distribution of the transposon insertions (data not shown). To further characterize the distribution of the Tn5367 insertions, the DNA sequences adjacent to the transposon insertions were determined for 11 kanr clones of BCG and 13 clones of M. tuberculosis (Table 5). All transposon insertions analyzed were accompanied by an 8-bp target duplication, as was described for nine other Tn5367 insertions (12). The total of 33 insertions is within different target sites, demonstrating that Tn5367 transposes with little or no sequence specificity. The chromosomal DNA sequence into which Tn5367 had inserted could be readily identified because they revealed 100% identity on a DNA level with reported cosmid sequences (28). The inactivated genes could be predicted after a blast analysis compared with the National Center for Biotechnology Information database (Table 5). Notably, the leuD auxotroph mc21500 isolated in BCG resulted from Tn5367 insertion in the sixth codon of the leuD gene. This insertion site is different from mc2797 leuD and mc2798 leuD transposon mutants reported previously (12). In two M. tuberculosis mutants, mc23014 and mc23019, two independent Tn5367 insertions at widely separated regions of a large gene that has homology to polyketide biosynthesis gene were identified. The insertions in BCG and M. tuberculosis have been positioned on the M. tuberculosis H37Rv physical map (28), and we show that the insertions are scattered within every quadrant of the genome (Fig. 3).
Table 4.
Effect of incubation time on transpositions in BCG and M. tuberculosis
| Strain | Time of adsorption, λ | kanr colonies/109 input cells
|
|
|---|---|---|---|
| phAE77 | phAE94 | ||
| BCG | 0.5 | 0 | 0 |
| BCG | 2.0 | 0.9 × 10−6 | 0 |
| BCG | 4.0 | 1.4 × 10−6 | 0 |
| BCG | 6.0 | 1.2 × 10−6 | 2 × 10−8 |
| BCG | 8.0 | 0.5 × 10−6 | 1 × 10−8 |
| M. tuberculosis (Erdman) | 6.0 | 5 × 10−8 | 1.7 × 10−7 |
| M. tuberculosis (Erdman) | 6.0 | 0 | 3.5 × 10−7 |
| M. tuberculosis (Erdman) | 6.0 | ND | 8.0 × 10−7 |
ND, not determined.
Table 5.
Mapping of Tn5367 insertion mutants in M. tuberculosis and BCG
| Mutant | Gene interrupted | SC or MC cosmid | Sequence of the transposon duplication |
|---|---|---|---|
| M. tuberculosis | Erdman | ||
| mc23002 | Unknown | cY75.unf (SC) | CATTCATT |
| mc23003 | Ferritin H (rsgA) | MTCY1A6 (SC) | AATAAACC |
| mc23004 | Unknown | cY23H3 (SC) | CGTTATCG |
| mc23005 | Unknown | cSCY03A11 (SC) | TGTTTGAC |
| mc23006 | Transketolase | cY454 (SC) | GTGAAACC |
| mc23007 | Unknown | MTCY31 (SC) | GCTTTTAC |
| mc23008 | Acetyl–CoA synthetase | MSGY409 (MS) | GGTTTTGA |
| mc23011 | Unknown | cY28.unf (SC) | GGTTTCCC |
| mc23014 | Polyketide synthase | cSCY22G10 (SC) | CAGTAACG |
| mc23015 | Thiosulfate sulfur transferase | cY164 (SC) | GGTGATCC |
| mc23017 | Imidazole glycerol phosphate dehydratase | Not found | GTGATACA |
| mc23019 | Polyketide synthase | cY275,unf (SC) | CGGTTTGC |
| mc23020 | Alcohol dehydrogenase (C terminus) | MTCY190 (SC) | TGGAAACC |
| BCG | |||
| mc21500 | leuD (insertion in the 6th codon) | Not found | GTGAAAGG |
| mc21501 | DR | cSCY16B7 (SC) | CCAAAACC |
| mc21502 | DR | cSCY16B7 (SC) | CAAAACCC |
| mc21503 | Unknown | Not found | CAAAACCC |
| mc21504 | Unknown | cY28.unf (SC) | TTCAAACT |
| mc21505 | Translation elongation factor G (efg) | Not found | GTTTTTCG |
| mc21506 | 8-Amino-7-oxononaate synthase (bioF) | MTCY10H4 (SC) | ACATTTGT |
Homology to predicted ORF or analogous functions by alignment (blast) to National Center for Biotechnology Information database. DR, direct repeat; SC, Sanger Center, U.K.; MC, Genomic Therapeutics Corporation (Waltham, MA).
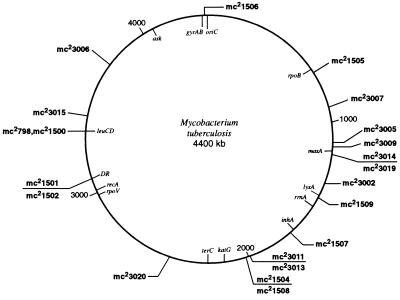
Figure 3.
Distribution of Tn5367 insertions on the genome map of M. tuberculosis. The location of the Tn5367 insertions in mutants generated from M. tuberculosis Erdman [mc2 3000s] or BCG [mc2 1500s] were deduced from the determination of the sequence adjacent to the insertion and comparison to the M. tuberculosis genomic database and the M. tuberculosis physical map (28).
DISCUSSION
To facilitate transposon-mediated insertional mutagenesis in pathogenic mycobacteria, we have developed conditionally replicating shuttle phasmids that efficiently deliver transposons to both fast-growing M. smegmatis and M. phlei species and the slow-growing species BCG and M. tuberculosis. Although, in the fast-growing mycobacteria, transposon mutagenesis has been achieved using nonreplicating plasmids (7–9, 12), phage delivery offers significant advantages for transposon mutagenesis of BCG and M. tuberculosis. The nonreplicating plasmids are of limited use in slow-growing mycobacteria because of the low transformation frequencies obtained with electroporation. The disadvantage of the conditionally replicating plasmid system is that, unless the transposition event is regulated, there is no means to select against propagation of transposon mutants during the outgrowth stage, with the consequence that many of the mutants selected will be siblings of a few mutated cells. In contrast, with a conditionally replicating phage system in which essentially every cell is infected, the selection can be executed shortly after infection, and the mutations are likely to represent independent events. Analysis of the BCG- and M. tuberculosis-independent mutants obtained in this study suggests that this is the case.
The transposon Tn5367 that we have used for insertional mutagenesis in this study has several particularly desirable features. As shown previously (12) and confirmed in this study, Tn5367 transposes in a relatively random fashion and therefore should allow for insertions in virtually every gene of a mycobacterial genome. Transposition frequencies could be improved by fusing the transposase to more active promoters or expression signals and additionally provide transposon elements (mini-Tn5367) that can no longer transpose once dissociated from their transposase. Transposon-containing, promoter-less reporter genes could be constructed that would allow for the generation of libraries that permit the screening of regulated promoters, as has been done in other genera (5).
In addition to delivering useful transposon constructs, the conditionally replicating shuttle phasmids represent potentially versatile vectors for genetic transfer. At present, we know that an additional 4 kb of DNA can be introduced into the pYUB552 backbone and still maintain the ability of the shuttle phasmids to yield functional mycobacteriophage. In addition to delivering transposons and creating marked insertional mutations, these vectors could be used to deliver any gene(s) to M. tuberculosis in a transient fashion. Infection of M. tuberculosis with conditionally replicating TM4 shuttle phasmid containing the firefly luciferase gene has already increased the sensitivity of the luciferase phage reporter assay (25). Infection at the nonpermissive temperature has been found both to increase the cumulative light emission and extend the time available for the assay because the phage is unable to proceed to lytic growth (25). Transient delivery of genes encoding γδ-resolvase or Cre recombinase to M. tuberculosis offers unique possibilities for precise gene deletions and removal of antibiotic selective markers that will be valuable in the creation of nonreverting, attenuated vaccine strains acceptable for use in humans. It is our hope that these phages may allow the development of specialized transducing phages that will efficiently mediate allelic exchange in M. tuberculosis.
The ability to generate libraries of transposon mutants in M. tuberculosis together with the complete genomic sequence shortly to become available provides an unprecedented opportunity to make rapid and substantial progress in the understanding of pathogenesis and development of novel therapeutics for tuberculosis. The rapid determination of sequences adjacent to the transposon insertion allows for an immediate classification of a gene as being nonessential for the multiplication of the tubercle bacillus. The set of essential genes would represent attractive candidates for analysis as practical drug targets. Many nonessential genes may represent important virulence determinants, as well. Indeed, among the mutants analyzed, it will be of immediate interest to examine the insertions in the mycocerosic acid biosynthetic gene (26) and the polyketide-biosynthetic gene [which shares significant homology to 6 of the 14 modules of the immunosuppressant rapamycin biosynthetic gene (27)] for their possible roles in virulence of M. tuberculosis. It is our view that mutational analyses represent the most powerful approach available to dissect the steps and functions governing the pathogenic process of M. tuberculosis and to support our hope that the conditionally replicating shuttle phasmids will be a valuable tool in facilitating that analysis.
Acknowledgments
We thank Marty Pavelka, Keith Derbyshire, and Jeffery Cox for their critical reading of this manuscript and Vladimir Pelicic, Christoph Guilhot, Mary Jackson, Jean-Mark Reyrat and Brigitte Gicquel for sharing their results before publication. This work was supported by grants from the National Institutes of Health (AI26170, AI27235, and AI23545) and the Burroughs Wellcome Fund.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BCG, Bacille Calmette–Guérin; kanr, kanamycin resistance; MOI, multiplicity of infection; IS, insertion sequence.
References
- 1.Murray C J L, Styblo K, Rouillon A. Bull Int Union Tuberc Lung Disorders. 1990;65:6–26. [PubMed] [Google Scholar]
- 2.Murray C J L, Lopez A D. Global Burden of Diseases. Cambridge, MA: Harvard Univ. Press; 1997. p. 273. [Google Scholar]
- 3.Tokunaga T, Mizuguchi Y, Suga K. J Bacteriol. 1973;113:1104–1111. doi: 10.1128/jb.113.3.1104-1111.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudaraj C V, Ramakrishnan T. Nature (London) 1971;228:280–281. [Google Scholar]
- 5.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 6.Berg C M, Berg D E, Groisman E A. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, D.C.: Am. Soc. Microbiol.; 1989. pp. 879–925. [Google Scholar]
- 7.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Nature (London) 1990;21:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 8.Fomukong N G, Dale J W. Gene. 1993;130:99–105. doi: 10.1016/0378-1119(93)90351-3. [DOI] [PubMed] [Google Scholar]
- 9.England P M, Wall Q, McFadden J. Mol Microbiol. 1991;5:2047–2052. doi: 10.1111/j.1365-2958.1991.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 10.Cirillo J D, Barletta R G, Bloom B R, Jacobs W R., Jr J Bacteriol. 1991;173:7772–7780. doi: 10.1128/jb.173.24.7772-7780.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalpana V G, Bloom B R, Jacobs W R., Jr Proc Natl Acad Sci USA. 1991;88:5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAdam R A, Weisbrod T R, Martin J, Scuderi J D, Brown A M, Cirillo J D, Bloom B R, Jacobs W R., Jr Infect Immunol. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balasubramanian V, Pavelka M S, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guilhot C, Otal I, Van Rompaey I, Martin C, Gicquel B. J Bacteriol. 1994;176:535–539. doi: 10.1128/jb.176.2.535-539.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs W R, Jr, Tuckman M, Bloom B R. Nature (London) 1987;327:532–536. doi: 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 16.Snapper S B, Lugosi L, Jekkel A, Melton R E, Kieser T, Bloom B R, Jacobs W R., Jr Proc Natl Acad Sci USA. 1988;85:6987–6991. doi: 10.1073/pnas.85.18.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs W R, Jr, Barletta R, Udani R, Chan J, Kalkut G, Sarkis G, Hatfull G F, Bloom B R. Science. 1993;260:819–822. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 18.Pearson R E, Jurgensen G J, Sarkis G J, Hatfull G F, Jacobs W R., Jr Gene. 1996;183:129–136. doi: 10.1016/s0378-1119(96)00530-6. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: ALaboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Pavelka M S, Jr, Jacobs W R., Jr J Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Udani R, Jones Jr W D, Barletta R, Bloom B R. Methods Enzymol. 1991;204:535–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 22.Davis R W, Botstein D, Roth J R. Advanced Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1980. pp. 94–95. [Google Scholar]
- 23.Snapper S B, Melton R E, Mustafa S, Kreser T, Jacobs W R., Jr Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 24.Telenti A, Marchesi F, Balz M, Bally F, Bottger E C, Bodmer T. J Clin Microbiol. 1973;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carriere, C., Riska, P., Oren, Z., Kriakov, J., Bardarov, S., Burns, J., Chen, J. & Jackobs, W. R., Jr. (1997) J. Clin. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 26.Azad A K, Sirakova T D, Rogers L M, Kollattukudy P E. Proc Natl Acad Sci USA. 1966;93:4787–4792. doi: 10.1073/pnas.93.10.4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwecke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillippe W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]