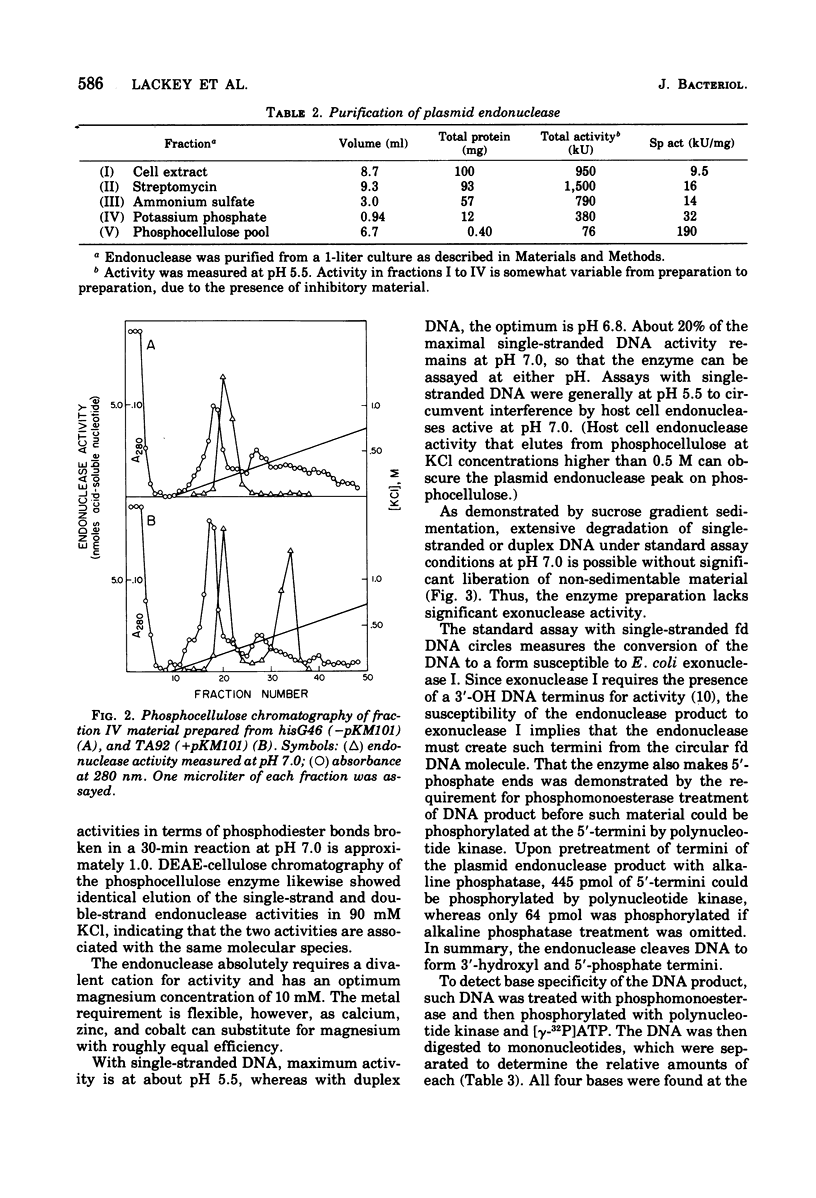
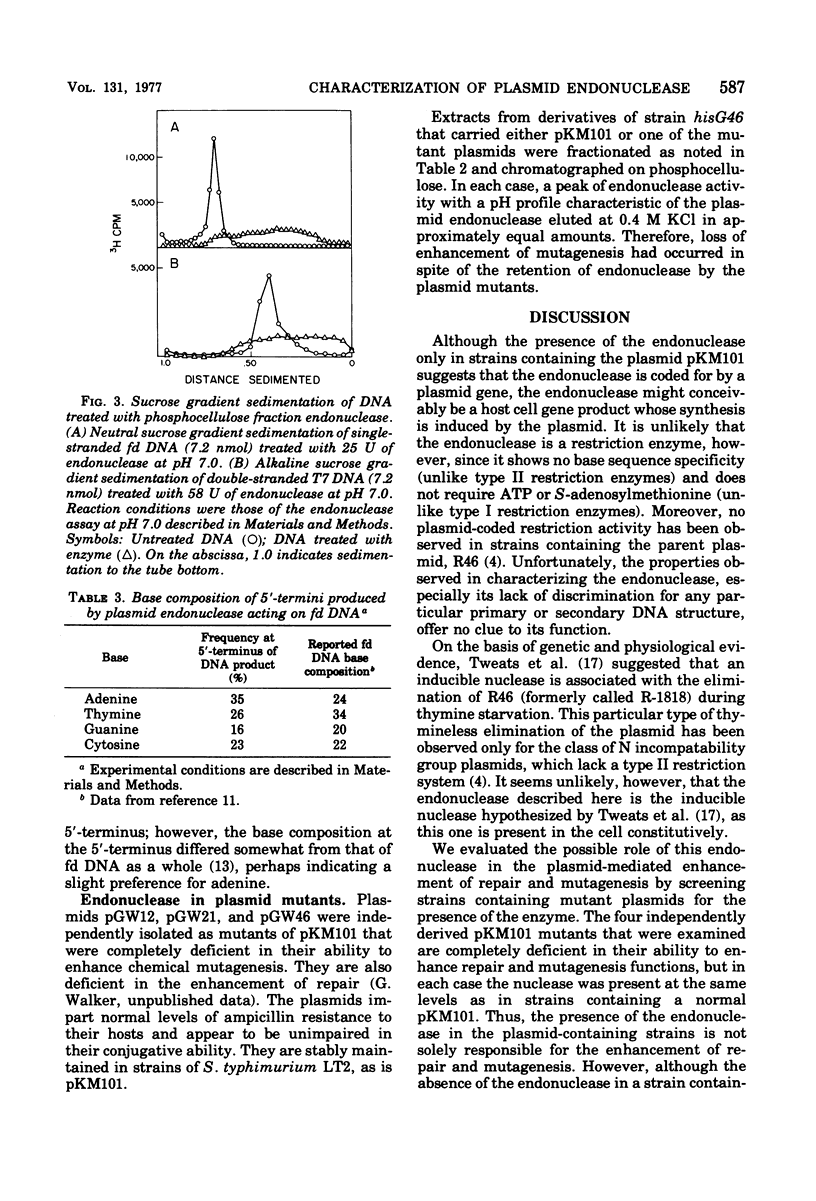
Abstract
An endonuclease was detected in strains of Salmonella typhimurium containing the drug resistance plasmid pKM101. The enzyme was not detectable in strains lacking this plasmid, but it was present in strains containing mutants of pKM101 that were no longer able to enhance host cell mutagenesis. The endonuclease had a molecular weight of roughly 75,000 and, at pH 7.0, was equally active on single-stranded and duplex deoxyribonucleic acid (DNA). The reaction with single-stranded DNA was optimal at pH 5.5, whereas with duplex DNA the optimum was pH 6.8. The enzyme required a divalent cation for activity, and it had no detectable exonuclease activity with single-stranded or duplex DNA. The endonuclease extensively degraded DNA with no apparent base specificity, forming 5'-phosphomonoester termini. Although characterization of the endonuclease has not revealed its function, the enzyme does not appear to be a restriction endonuclease.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alper M. D., Ames B. N. Positive selection of mutants with deletions of the gal-chl region of the Salmonella chromosome as a screening procedure for mutagens that cause deletions. J Bacteriol. 1975 Jan;121(1):259–266. doi: 10.1128/jb.121.1.259-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B. N., Lee F. D., Durston W. E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc Natl Acad Sci U S A. 1973 Mar;70(3):782–786. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B. N., Mccann J., Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975 Dec;31(6):347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Birks J. H., Pinney R. J. Correlation between thymineless elimination and absence of hsp II (EcoRII) specificity in N-group R factors. J Bacteriol. 1975 Mar;121(3):1208–1210. doi: 10.1128/jb.121.3.1208-1210.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark P. J., Linn S. An endonuclease activity from Escherichia coli absent from certain rec- strains. Proc Natl Acad Sci U S A. 1970 Sep;67(1):434–441. doi: 10.1073/pnas.67.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark P. J., Linn S. Purification and properties of the recBC DNase of Escherichia coli K-12. J Biol Chem. 1972 Mar 25;247(6):1849–1860. [PubMed] [Google Scholar]
- Jacquemin-Sablon A., Richardson C. C. Analysis of the interruptions in bacteriophage T5 DNA. J Mol Biol. 1970 Feb 14;47(3):477–493. doi: 10.1016/0022-2836(70)90316-5. [DOI] [PubMed] [Google Scholar]
- Kuhnlein U., Penhoet E. E., Linn S. An altered apurinic DNA endonuclease activity in group A and group D xeroderma pigmentosum fibroblasts. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1169–1173. doi: 10.1073/pnas.73.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEHMAN I. R., NUSSBAUM A. L. THE DEOXYRIBONUCLEASES OF ESCHERICHIA COLI. V. ON THE SPECIFICITY OF EXONUCLEASE I (PHOSPHODIESTERASE). J Biol Chem. 1964 Aug;239:2628–2636. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Marvin D. A., Hohn B. Filamentous bacterial viruses. Bacteriol Rev. 1969 Jun;33(2):172–209. doi: 10.1128/br.33.2.172-209.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortelmans K. E., Stocker B. A. Ultraviolet light protection, enhancement of ultraviolet light mutagenesis, and mutator effect of plasmid R46 in Salmonella typhimurium. J Bacteriol. 1976 Oct;128(1):271–282. doi: 10.1128/jb.128.1.271-282.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. The 5'-terminal nucleotides of T7 bacteriophage deoxyribonucleic acid. J Mol Biol. 1966 Jan;15(1):49–61. doi: 10.1016/s0022-2836(66)80208-5. [DOI] [PubMed] [Google Scholar]
- Tweats D. J., Pinney R. J., Smith J. T. R-factor-mediated nuclease activity involved in thymineless elimination. J Bacteriol. 1974 Jun;118(3):790–795. doi: 10.1128/jb.118.3.790-795.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Plasmid (pKM101)-mediated enhancement of repair and mutagenesis: dependence on chromosomal genes in Escherichia coli K-12. Mol Gen Genet. 1977 Mar 28;152(1):93–103. doi: 10.1007/BF00264945. [DOI] [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]



