Abstract
The host response to Gram-negative bacterial infection is influenced by two homologous lipopolysaccharide (LPS)-interactive proteins, LPS-binding protein (LBP) and the bacteridical/permeability-increasing protein (BPI). Both proteins bind LPS via their N-terminal domains but produce profoundly different effects: BPI and a bioactive N-terminal fragment BPI-21 exert a selective and potent antibacterial effect upon Gram-negative bacteria and suppress LPS bioactivity whereas LBP is not toxic toward Gram-negative bacteria and potentiates LPS bioactivity. The latter effect of LBP requires the C-terminal domain for delivery of LPS to CD14, so we postulated that the C-terminal region of BPI may serve a similar delivery function but to distinct targets. LBP, holoBPI, BPI-21, and LBP/BPI chimeras were compared for their ability to promote uptake by human phagocytes of an encapsulated, phagocytosis-resistant strain of Escherichia coli. We show that only bacteria preincubated with holoBPI are ingested by neutrophils and monocytes. These findings suggest that, when extracellular holoBPI is bound via its N-terminal domain to Gram-negative bacteria, the C-terminal domain promotes bacterial attachment to neutrophils and monocytes, leading to phagocytosis. Therefore, analogous to the role of the C-terminal domain of LBP in delivery of LPS to CD14, the C-terminal domain of BPI may fulfill a similar function in BPI-specific disposal pathways for Gram-negative bacteria.
Lipopolysaccharide (LPS)-binding protein (LBP) and bacteridical/permeability-increasing (BPI) protein are both 456-residue LPS-binding proteins (1). LBP is a plasma protein produced by the liver constitutively and in increased amounts during the acute phase response (2). The strongly cationic BPI is a constituent of polymorphonuclear leukocytes (PMN), stored in primary granules (3, 4). Despite the similar size, ≈45% sequence identity, and other structural similarities, the biological roles of LBP and BPI as LPS-complexing proteins are those of agonist and antagonist. Whereas LBP functions prominently in alerting the host to the presence of LPS by promoting delivery of LPS to cellular response systems (2), BPI suppresses LPS bioactivity and is a potent antibacterial agent with selective toxicity toward Gram-negative bacteria (3).
The determinants of LPS binding in the LBP molecule are located in its N-terminal half (residues 1–197) whereas the C-terminal half is needed for delivery of LPS to CD14, an important component in the LPS-signaling cascade (2, 5, 6). Thus, LBP is structurally and functionally organized as a two-domain molecule. This may also be the case for BPI, which can be cleaved into two nearly equally sized fragments by limited proteolysis (7, 8). The antibacterial and LPS-neutralizing activities of BPI are fully expressed by its N-terminal half (residues 1–193) (7, 9–11); however, the function of the C-terminal portion has remained largely undefined.
Notwithstanding their contrasting functions, the similarities between the two proteins raised the possibility that the C-terminal half of BPI might also serve a delivery function as part of the role of holoBPI in the disposal of Gram-negative bacteria. The demonstration that potently antibacterial concentrations of BPI accumulate extracellularly during inflammation (12) led us to speculate that BPI might have opsonic properties requiring the C-terminal half of the molecule. This hypothesis was tested by comparing the uptake by peripheral blood leukocytes of encapsulated, phagocytosis-resistant Escherichia coli preincubated with either holoBPI, a bioactive N-terminal fragment of BPI (BPI-21), holoLBP, and two chimeric constructs (BPI/LBP and LBP/BPI). The results show that, of these proteins, only holoBPI has opsonic effects, indicating that both the N-terminal and C-terminal portions of BPI are needed for opsonophagocytosis. Thus, BPI and LBP indeed share a two-domain structure with the C-terminal half of each protein serving a delivery function but to different targets.
MATERIALS AND METHODS
Bacteria.
E. coli K1/r, a K1-encapsulated strain with a rough LPS chemotype (12), was grown in 0.8% nutrient broth (Difco) in physiological saline. Overnight cultures were diluted 1:50 into fresh medium and grown to late log phase (≈4 h) at 37°C with vigorous shaking. Bacteria were sedimented at 3000 × g for 10 min and resuspended to 5 × 107/ml in sterile Hanks’ balanced salt solution without phenol red or divalent cations [Hanks’(−); BioWhittaker], buffered with 10 mM pyrogen-free Hepes (pH 7.4)(Sigma). Bacterial concentrations were determined by A550 using a Beckman DU-30 spectrophotometer.
LPS.
Purified LPS from E. coli J5 (Rc chemotype; Sigma) was resuspended to 1 mg/ml by sonication and vigorous vortexing in sterile pyrogen-free water and was stored at 4°C. Immediately before use, solutions were sonicated for 2 min at room temperature in a water bath sonicator (Sonic Dismembranator 550; Fisher Scientific) at 30% maximal output. LPS was serially diluted in Hanks’(−)/10 mM Hepes/1% (wt/vol) human serum albumin (HSA; Baxter Health Care, Mundelein, IL) with vigorous vortexing.
Proteins.
Native human BPI (holoBPI) was purified from PMN as described (9, 13). Recombinant human LBP, holoBPI, a bioactive N-terminal fragment of BPI (comprising residues 1–193; BPI-21) and two chimeric proteins consisting of either the N-terminal region of BPI (residues 1–200) fused to the C-terminal region of LBP (residues 199–456; BPI/LBP) or the N-terminal region of LBP (residues 1–197) fused to the C-terminal region of BPI (residues 200–456; LBP/BPI) were provided by Stephen Carroll (Xoma Corporation, Berkeley, CA). The design and purification of the recombinant BPI/LBP chimeras have been described (14). Antibodies used included a mouse mAb directed against an epitope within the N-terminal region of BPI (mAb 9–3/5), affinity-purified polyclonal rabbit anti-human LBP antibodies (provided by Xoma Corporation), and polyclonal rabbit anti-human BPI serum (15). A mAb (MY4) directed against CD14 was obtained from Coulter, and a mAb (clone 44) directed against the CD11b chain of complement receptor 3 (CR3) was a generous gift of M. Amin Arnout (Department of Medicine, Massachusetts General Hospital, Boston, MA). Antibodies (sera) were diluted in Hanks’(−)/10 mM Hepes/1% HSA [Hanks’(−)/Hepes/HSA].
Isolation and Preparation of Human Leukocytes.
Venous blood was drawn from healthy human volunteers (after informed consent) into a heparinized syringe, and leukocytes were isolated by sedimentation in 3% dextran (pyrogen-free, United States Biochemical). The platelet- and leukocyte-rich upper phase was spun at ≈30 × g for 10 min at room temperature in a swinging bucket rotor to sediment the leukocytes. For the opsonophagocytosis assays, erythrocytes were removed by hypotonic lysis, and the leukocytes were diluted to 1.7 × 107/ml in 10 mM Hepes-buffered (pH 7.4) Hanks’(−) (to prevent cell aggregation). To determine the role of mCD14 or CR3 in phagocytosis, the leukocytes were preincubated for 15 min at 37°C with 10 μg/ml of mAb to CD14, CR3, or buffer and then diluted 10× into reaction mixtures containing bacteria. For the signaling assays, the leukocytes were washed twice with 15 ml of Hanks’(−)/Hepes and resuspended to 107/ml.
Opsonophagocytosis and Viability.
Bacteria (2.5 × 107/ml) were preincubated for 30 min at 37°C with gentle shaking either alone or with 10, 30, or 90 nM protein or 5% pooled human serum in Hanks’(−)/Hepes (pH 7.4). After this incubation, an aliquot was taken, serially diluted in sterile physiological saline, and then plated (10 μl) in 5 ml of molten (48°C) 1.5% (wt/vol) agar (Difco) containing 0.8% nutrient broth in physiological saline. Bacterial colonies were counted after incubation at 37°C for 18–24 h. Leukocytes were added to the remaining reaction mixture for a final concentration of 2 × 106/ml and a final bacteria-to-leukocyte ratio of 10:1. Ca2+ and Mg2+ were added to a final concentration of 1.26 and 0.9 mM, respectively. Incubations were for 30 min at 37°C with gentle shaking followed by 4× dilution with cold trypticase soy broth (Beckton Dickinson Microbiology Systems). An aliquot of the diluted cell suspension (0.1 ml) was spun onto glass slides for 5 min at 500 rpm using a Shandon Cytospin 2 (Pittsburgh). The slides were air-dried for 10 min and stained with Wright–Giemsa (Fisher Scientific) according to the manufacturer’s instructions. The cells were visualized by light microscopy (Labophot, Nikon, Japan), and the number of apparently intracellular bacteria/100 PMN or monocytes was counted.
Transmission Electron Microscopy.
E. coli were preincubated with 90 nM holoBPI for 30 min and then incubated with leukocytes for 30 min as described above. The leukocytes then were sedimented and fixed for 30 min at room temperature in 2% glutaraldehyde in 0.1 M Na cacodylate buffer (pH 7.4). The cells were washed once and incubated overnight in the same fixative. The cells were washed 3 times in cacodylate buffer, postfixed in 2% OsO4/cacodylate buffer for 1 h, and washed as above. The sample was stained with 1% uranyl acetate for 20 min, dehydrated with graded alcohols, embedded in epoxy resin, and polymerized over 5 days at 68°C. Thin sections of ≈60–80 nm were stained with uranyl acetate and lead citrate, washed, dried, and observed in a JEOL JEM-1200 EX II electron microscope.
SDS/PAGE and Immunoblotting.
SDS/PAGE was carried out in 10–15% gradient gels run on the Phast-Gel System (Pharmacia) according to the manufacturer’s instructions after heating the samples for 10 min in a boiling water bath. Immunoblotting onto a 0.45-μm nitrocellulose membrane (Schleicher & Schuell, Keene, NH) was carried out as described (16) except that the transfer was allowed to proceed overnight at 4°C at 200 mAmps. After transfer, the gels were stained with Coomassie brilliant blue (Bio-Rad) to confirm transfer of protein from the gels. The nitrocellulose blots were blocked at 37°C for 1 h in PBS (pH 7.4) containing 0.05% polyoxyethylene sorbitan monolaurate (Tween-20; Bio-Rad) and 3% gelatin (EM Science), washed 3 times in PBS containing 0.05% Tween, and incubated for 3 h at 37°C with one of the following antibodies diluted 1000× in PBS containing 0.05% Tween and 1% gelatin: rabbit polyclonal anti-BPI for detection of BPI-21; mouse monoclonal anti-BPI for detection of holoBPI, or rabbit polyclonal anti-LBP for detection of BPI/LBP, LBP/BPI, and LBP. The blots were washed as before, and bound IgG was detected by a 2- to 3-h incubation of 0.05 μCi/ml of I125-Protein G (12 mCi/mg; Amersham) diluted in PBS containing 0.05% Tween and 1% gelatin. The blots were washed, air dried, and exposed to Kodak X-Omat film for 5–7 days.
Chemiluminescence.
Bacteria (1.25 × 107/ml) or LPS (125 ng/ml) were preincubated alone or with 112.5 nM protein (holoBPI, BPI-21, BPI/LBP, LBP/BPI, or LBP) for 30 min at 37°C in Hanks’(−)/Hepes/HSA. After this preincubation, aliquots (80 μl) of this mixture were added undiluted or diluted 10-, 100-, or 1000-fold in Hanks’(+)/Hepes/HSA to 96-well plates (Optiplate, Packard) containing leukocytes in Hanks’(+)/Hepes/HSA and either lucigenin (bis-N-methylacridinium nitrate) or luminol (3-aminophthalhydrazide; Sigma). The final reaction mixture contained 106 leukocytes/ml and 0.1, 1, 10, or 100 ng LPS per ml or 104, 105, 106, or 107 bacteria/ml ± 0.09, 0.9, 9, or 90, nM various BPI or LBP species and lucigenin or luminol (30 μM either) in a final volume of 100 μl. Chemiluminescence was measured at 37°C with 5 s of gentle shaking every 5 min in an Anthos Lucy I luminometer (Columbia Bioscience, Frederick, MD). Data were collected for 5 s/sample at 10-min intervals for 2 h. In the absence of bacteria or LPS, leukocyte chemiluminescence (plus or minus added proteins) was negligible (<100 photons/s).
RESULTS AND DISCUSSION
HoloBPI Promotes Phagocytosis of E. coli K1/r by Human Neutrophils.
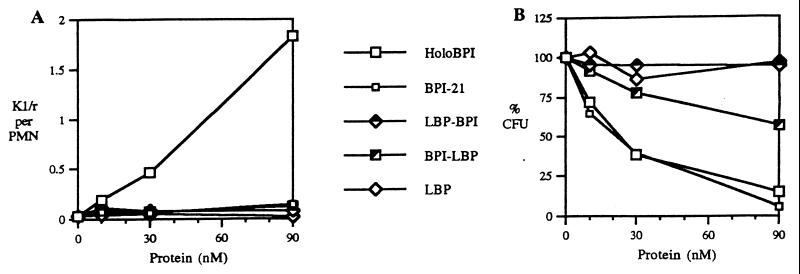
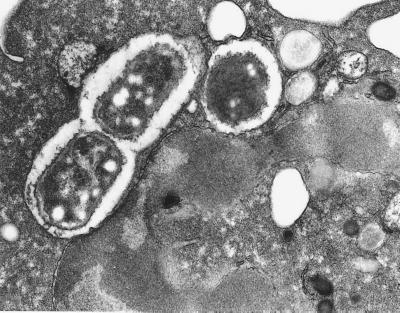
The extracellular accumulation of BPI in PMN-rich exudates (12) and the role of the structurally related LBP in delivery of LPS and Gram-negative bacteria to mCD14 on myeloid cells (2) suggested that BPI could also play a role in delivery of bacteria to phagocytic cells. To test this possibility, we determined the effect of BPI on uptake by leukocytes of E. coli K1/r, a BPI-sensitive encapsulated strain that is resistant to phagocytosis in the absence of opsonins (Fig. 1A; <0.1 bacteria ingested/PMN, <1% of the added bacteria ingested). BPI in inflammatory fluids displays potent antibacterial activity toward E. coli K1/r (12), reflecting avid binding to these encapsulated bacteria (C. Capodici and J.W., unpublished observations). However, because such biological fluids contain other potentially opsonic and/or antibacterial factors (12, 17), E. coli K1/r were pretreated with BPI in an artificial medium (balanced salts solution) to assess unambiguously whether BPI has an opsonic effect. Addition of BPI promoted bacterial uptake in a dose-dependent fashion; significant uptake was manifest with as little as 10 nM holoBPI (Fig. 1A). Native and recombinant holoBPI had equivalent opsonic activity toward E. coli K1/r and J5, a nonencapsulated rough strain (data not shown). Uptake of bacteria in the presence of holoBPI was 6× greater by PMN than by monocytes whereas bacteria opsonized with 5% pooled human serum were taken up equally by monocytes and PMN (Table 1). The intracellular location of PMN-associated bacteria opsonized by BPI was confirmed by electron microscopy (Fig. 2).
Figure 1.
Comparison of effect of increasing concentrations of various BPI and/or LBP species on (A) uptake of E. coli K1/r and (B) viability of bacteria. Bacterial uptake is expressed as number of PMN-associated E. coli/PMN (at least 100 PMN counted/sample). Bacterial viability is expressed as a percentage of viability of bacteria incubated alone. All data shown represent the mean of the same three experiments.
Table 1.
BPI-mediated phagocytosis is independent of mCD14 and CR3
| Cell type and antibody | Opsonin
|
||
|---|---|---|---|
| HoloBPI | Serum | None | |
| Neutrophils | 100.0 | 63.4 | 1.6 |
| +MY4 | 105.6 | 88.1 | 3.5 |
| +αCR3 | 100.7 | 21.7 | 2.1 |
| Monocytes | 16.4 | 65.6 | 0.0 |
| +MY4 | 28.0 | 69.9 | 0.7 |
| +αCR3 | 28.0 | 35.0 | 1.0 |
Bacterial uptake is expressed as a percentage of uptake by PMN of BPI-treated bacteria (=100%). The data shown represent the mean of two experiments.
Figure 2.
Thin section electron micrograph of a single PMN with three bacteria visible within two phagocytic vacuoles. E. coli were preincubated with 90 nM holoBPI before incubating with leukocytes and processing for electron microscopy, as described in Materials and Methods.
Role of N- and C-terminal Regions of BPI in Opsonophagocytosis of E. coli K1/r.
LBP-mediated delivery of LPS to CD14 depends on the combined functions of the N- and C-terminal halves of the protein in, respectively, binding of LPS and delivering LPS to CD14 (5, 6). To determine whether the opsonic effects of BPI similarly depend on distinct but interdependent functions of its N- and C-terminal domains, we compared the ability of various BPI and LBP species to promote uptake of E. coli K1/r by leukocytes. BPI-21, a recombinant protein species corresponding to the bioactive N-terminal domain of BPI, contains all of the described bioactivities of holoBPI, including bactericidal activity and LPS neutralization (9–11). As expected, holoBPI and BPI-21 produced the same dose-dependent growth inhibition of E. coli K1/r (Fig. 1B). However, in contrast to holoBPI, BPI-21 did not promote uptake of E. coli K1/r even when up to 10× the minimal effective holoBPI dose was used (Fig. 1A) and incubations were extended from 30 to 60 min. These data suggest a requirement for the C-terminal region of BPI to mediate opsonophagocytosis of E. coli K1/r. Consistent with this interpretation, neither LBP nor BPI/LBP promoted phagocytosis of E. coli K1/r (Fig. 1A). In the dose range tested, LBP/BPI was also unable to promote uptake of K1/r, suggesting that both the N- and C-terminal domains derived from BPI were needed for opsonic activity toward K1/r. This finding may reflect the lower affinity of the N-terminal region of LBP for LPS (18, 19), producing insufficient bacterial binding for the BPI-derived C-terminal region to mediate an opsonic effect.
The Affinity of BPI and LBP Species for E. coli K1/r Reflects the Origin of the N-Terminal Region of the Protein.
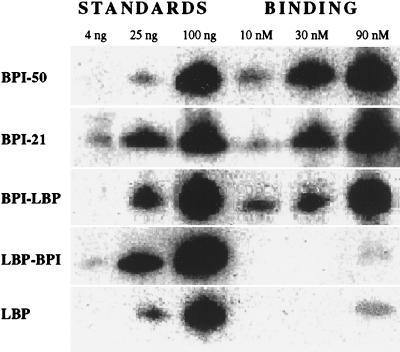
To determine directly the role of the N-terminal domain of BPI and LBP in binding to bacteria, protein binding to E. coli K1/r was assayed by Western blotting. Binding of holoBPI, BPI-21, and BPI/LBP was closely similar and ≥10× greater than that of LBP and LBP/BPI (Fig. 3). Thus, as demonstrated for binding to purified LPS (18, 19), we now show that binding to E. coli also is determined by the N-terminal regions of both BPI and LBP and that the affinity of this region of BPI is much higher than that of LBP. The similar bacterial binding of holoBPI and BPI-21 confirms that the difference in opsonic activity reflects an additional function of holoBPI, reflecting interactions between BPI-coated bacteria and phagocytes (PMN) involving the C-terminal half of BPI. The lack of opsonic activity of LBP/BPI is consistent with the weaker bacterial binding of this protein chimera. We attribute the finding that only holoBPI has opsonic activity to the concerted actions of the two halves of BPI; the N-terminal region of BPI attaching with high affinity to the target bacteria and the C-terminal region mediating interactions with phagocytes.
Figure 3.
Comparison of binding of various BPI and LBP species to E. coli K1/r. The bacteria were incubated with the indicated concentrations of the various proteins under the same conditions as Fig. 1. Bound protein (associated with 8 × 106 bacteria) was detected by immunoblotting, as described in Materials and Methods. Known amounts (4, 25, and 100 ng) of each protein were run on the same immunoblot as standards to facilitate quantitation of bound protein. Data shown are representative of at least two independent experiments.
BPI-Mediated Phagocytosis Does Not Depend on CD14 or CR3.
The membrane receptors CD14 and CR3 are present on both PMN and monocytes and play a role in attachment and internalization of Gram-negative bacteria (20–22). To determine the possible role of these proteins in BPI-mediated phagocytosis, we measured bacterial uptake in the absence or presence of neutralizing mAb to CD14 or CR3. Anti-CR3 substantially inhibited uptake of serum-opsonized E. coli K1/r but had no effect on BPI-mediated uptake (Table 1). Anti-CD14 nearly completely inhibited LPS-induced activation of the leukocytes (data not shown) but had no effect on either serum or BPI-mediated phagocytosis. Thus, BPI-mediated phagocytosis is independent of CR3 and CD14.
Opposite Effects of BPI and LBP on E. coli K1/r-Induced Leukocyte Activation: Role of the C-Terminal Region.
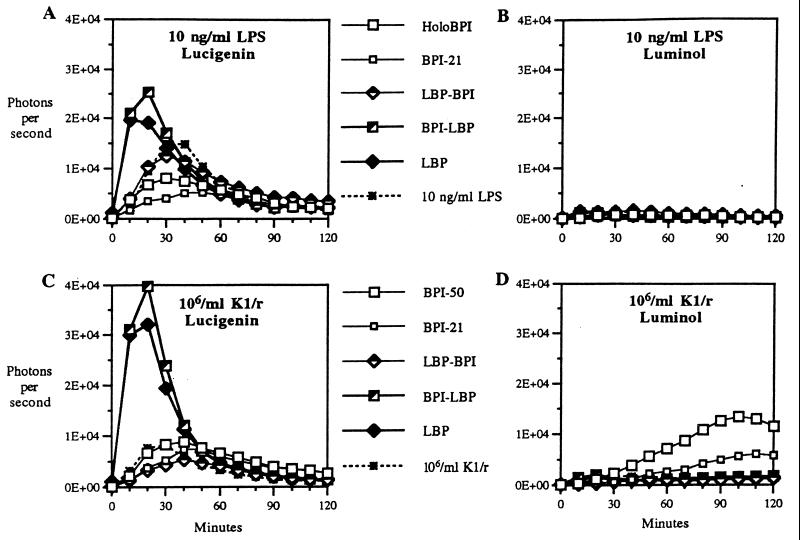
All previous studies have shown that LBP potentiates cellular activation induced by purified LPS or bacteria-associated LPS whereas BPI potently inhibits such signaling (2, 3, 11, 23, 24). Accordingly, BPI/LBP chimeras containing the C-terminal half of LBP promote LPS signaling whereas protein species containing the C-terminal half of BPI inhibit signaling, indicating an essential role of the C-terminal half of LBP in delivery of LPS to mCD14 and resulting cellular activation (14). The finding that BPI promotes uptake of intact bacteria by (LPS-responsive) leukocytes raised the question: Does BPI-mediated bacterial uptake trigger cellular activation? To test this possibility, we measured bacterial or purified LPS-induced leukocyte chemiluminescence, which sensitively monitors activation of the respiratory burst (25). Chemiluminescence was measured in the presence of lucigenin or luminol to distinguish accumulation of the primary products of the respiratory burst (e.g., O2−, H2O2; reactive with lucigenin) from oxidants derived from myeloperoxidase-mediated metabolism that are preferentially reactive with luminol (25). In the absence of BPI or LBP, both purified LPS and E. coli K1/r caused a dose-dependent (data not shown; 24) activation of leukocyte chemiluminescence that was apparent with lucigenin, but not with luminol, reflecting activation of the respiratory burst without concomitant mobilization by degranulation of myeloperoxidase (Fig. 4). Lucigenin-enhanced chemiluminescence [induced by both purified LPS (10 ng/ml) and E. coli (106/ml)] was stimulated by LBP and BPI/LBP but inhibited by BPI-21, holoBPI, and LBP/BPI (Fig. 4 A and C).§ In contrast, luminol-enhanced chemiluminescence was triggered by E. coli K1/r (but not by purified LPS) after treatment with holoBPI and to a lesser extent BPI-21 but not by the other proteins (Fig. 4 B and D). The effects of the various proteins on lucigenin-enhanced chemiluminescence are consistent with an essential role of the C-terminal domain of LBP in promoting LPS signaling mediated either by purified endotoxin or by intact E. coli in the absence of phagocytosis (24). The ability of BPI-21 to promote luminol-enhanced chemiluminescence suggests that alterations of the bacterial surface mediated by the antibacterial N-terminal domain can promote bacteria–phagocyte interactions that trigger degranulation even in the absence of phagocytosis. The much more pronounced stimulation of luminol-enhanced chemiluminescence by bacteria treated with holoBPI probably reflects the combined effects of bacterial surface alterations and ingestion of the bacteria. The inability of BPI/LBP to produce the same effect as BPI-21 is consistent with the (unexplained) lower antibacterial activity of this chimera (Fig. 1B) and its lack of opsonic activity (Fig. 1A). Mobilization of myeloperoxidase-mediated oxidative metabolism by BPI-coated bacteria may further enhance bacterial destruction by combining the action of PMN-derived nonoxidative and oxidative antibacterial systems. LPS aggregates treated with BPI do not evoke the same response (Fig. 4B), revealing a striking difference in the nature and/or consequences of BPI-mediated leukocyte interaction with bacteria vs. purified LPS. This difference may provide a means for restricting mobilization of potent but nonselective oxidative cytotoxicity to sites/compartments in close juxtaposition to target bacteria.
Figure 4.
Effect of the various BPI and/or LBP species on the triggering of lucigenin- (A and C) or luminol (B and D)-enhanced leukocyte chemiluminescence by purified LPS (A and B) or E. coli K1/r (C and D). Bacteria or LPS were preincubated plus or minus the indicated protein for 30 min before addition to leukocytes plus lucigenin or luminol and measurement of chemiluminescence as described in Materials and Methods. The results shown represent the mean of four determinations using data obtained during incubation of 10 ng/ml LPS or 106/ml E. coli with leukocytes.
These findings reveal a (opsonic) function of BPI. Unlike the previously described activities of BPI (cytotoxicity toward Gram-negative bacteria, endotoxin neutralization), opsonic activity requires both the N- and C-terminal halves of BPI, providing a bridge between Gram-negative bacteria and phagocytes. This function is analogous to the role of LBP in delivering LPS to myeloid cells containing mCD14, further illustrating how similar the organization of structure and function is in these two LPS-binding proteins. Recent x-ray structural analysis of BPI has revealed a highly elongated molecule with the opposite ends of the N- and C-terminal domains up to ≈120 Å apart (26). This structural configuration seems compatible with simultaneous binding of two different cells. Accordingly, sites enriched in basic residues that mediate the primary interactions of BPI with LPS (3) are concentrated within the N-terminal domain near one end of the molecule (27). Although the sites and properties within the C-terminal domain important for opsonic function are unknown, one prominent feature of this region of BPI is its hydrophobicity (9, 28). Because maximal opsonic effects are associated with binding of ≥100,000 molecules of BPI–bacterium (Figs. 1A and 3), the presence of the C-terminal domain on the bacterial envelope may increase bacterial surface hydrophobicity and thereby promote phagocytosis (29). Alternatively, more specific “ligand–receptor” interactions involving the C-terminal domain of BPI and as yet undefined phagocyte membrane receptors may contribute to phagocytosis in a manner analogous to the LPS/LBP interaction with mCD14.
Whatever the precise mechanism of BPI-mediated phagocytosis, these findings reveal an additional means by which BPI can contribute to host antibacterial defenses at inflammatory sites. Whereas our earlier studies have implicated a role for granule-associated BPI in intracellular killing of Gram-negative bacteria by PMN (30–32) and have revealed a potential role for secreted BPI in extracellular killing of phagocytosis-resistant encapsulated E. coli (12) and in regulation of Gram-negative bacterial (LPS)-mediated signaling (11, 24), we now propose a role for BPI in disposal of Gram-negative bacteria via ingestion by phagocytes. Although the opsonic effects described in this study required BPI doses that produced antibacterial (growth-inhibitory) effects (compare Fig. 1 A and B), it is possible that, in the presence of other opsonins and bacteria with different envelope properties, lower BPI concentrations may be sufficient for opsonization without direct cytotoxicity. In any case, the delivery of bacteria to phagocytes will promote bacterial clearance and intracellular disassembly and, hence, contribute further to the ultimate elimination of bacteria and bacterial products.
Acknowledgments
We thank Ivonna Gumper for her invaluable help with electron microscopy and David Sabatini for making his facility available for our use. We also thank Stephen Carroll for making available recombinant BPI, LBP species, M. Amin Arnout for generously providing the mAb to CR3, and all of the members of the lab, especially Yvette Weinrauch, Seth Katz, Ofer Levy, Kol Zarember, and Shu Chen, for their tireless advice-giving and assistance.
This work was supported in part by Public Health Service grant R37 DK 05472 as well as by a grant from the XOMA Corporation (Berkeley, CA). N.I. also was supported by National Institutes of Health Training Grant 5T32GM07308 from the National Institute of General Medical Science.
ABBREVIATIONS
- BPI
bactericidal/permeability-increasing protein
- Hanks’(−)
Hanks’ balanced salt solution without phenol red and divalent cations
- Hanks’(+)
Hanks’(−) with divalent cations
- HSA
human serum albumin
- LPS
lipopolysaccharide
- LBP
LPS-binding protein
- PMN
polymorphonuclear leukocyte
Footnotes
These conditions provide the most striking demonstration of the contrasting effects of the various proteins on lucigenin- vs. luminol-enhanced chemiluminescence. The stimulatory effects of LBP and BPI/LBP and the inhibitory effects of holoBPI, BPI-21, and LBP/BPI on lucigenin-enhanced chemiluminescence were greater when the stimulating agents were further diluted (to 1 ng/ml LPS and 105 E. coli/ml) before addition to leukocytes, but these conditions were too dilute for detection of effects on luminol-enhanced chemiluminescence. Conversely, the stimulatory effect of holoBPI on luminol-enhanced chemiluminescence was greater and occurred earlier (coincident with phagocytosis) at 107 bacteria/ml, but the effects of the proteins on lucigenin-enhanced chemiluminescence were dampened at such high bacteria/LPS concentrations.
References
- 1.Schumann R R, Leong S R, Flaggs G W, Gray P W, Wright S D, Mathison J C, Tobias P S, Ulevitch R J. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 2.Ulevitch R J, Tobias P S. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 3.Elsbach P, Weiss J. Immunobiology. 1993;187:417–429. doi: 10.1016/S0171-2985(11)80354-2. [DOI] [PubMed] [Google Scholar]
- 4.Zarember K, Elsbach P, Kim K-S, Weiss J. Blood. 1997;89:672–679. [PubMed] [Google Scholar]
- 5.Han J, Mathison J C, Ulevitch R J, Tobias P S. J Biol Chem. 1994;269:8172–8175. [PubMed] [Google Scholar]
- 6.Theofan G, Horwitz A G, Williams R E, Liu P S, Chan I, Birr C, Carroll S F, Meszaros K, Prent J B, Kasler H, Aberle S, Trown P W, Gazzano-Santoro H. J Immunol. 1994;152:3623–3629. [PubMed] [Google Scholar]
- 7.Ooi C E, Weiss J, Elsbach P, Frangione B, Mannion B A. J Biol Chem. 1987;262:14891–14894. [PubMed] [Google Scholar]
- 8.Capodici C, Weiss J. J Immunol. 1996;156:4789–4796. [PubMed] [Google Scholar]
- 9.Ooi C E, Weiss J, Doerfler M E, Elsbach P. J Exp Med. 1991;174:649–655. doi: 10.1084/jem.174.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gazzano-Santoro H, Parent J B, Grinna L, Horwitz A, Parsons T, Theofan G, Elsbach P, Weiss J, Conlon P J. Infect Immunol. 1992;60:4754–4761. doi: 10.1128/iai.60.11.4754-4761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss J, Elsbach O, Shu C, Castillo J, Grinna L, Horwitz A, Theofan G. J Clin Invest. 1992;90:1122–1130. doi: 10.1172/JCI115930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinrauch Y, Foreman A, Shu C, Zarember K, Levy O, Elsbach P, Weiss J. J Clin Invest. 1995;95:1916–1924. doi: 10.1172/JCI117873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannion B A, Kalatzis E S, Weiss J, Elsbach P. J Immunol. 1989;142:2807–2812. [PubMed] [Google Scholar]
- 14.Abrahamson S L, Wu H-M, Williams R E, Der K, Ottah N, Little R, Gazzano-Santoro H, Theofan G, Bauer R, Leigh S, Orme A, Horwitz A H, Carroll S F, Dedrick R L. J Biol Chem. 1997;272:2149–2155. doi: 10.1074/jbc.272.4.2149. [DOI] [PubMed] [Google Scholar]
- 15.Weersink A J L, Van Kessel K P M, van den Tol M E, van Strijp J A G, Torensma R, Verhoef J, Elsbach P, Weiss J. J Immunol. 1993;150:253–263. [PubMed] [Google Scholar]
- 16.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinrauch Y, Elsbach P, Madsen L M, Foreman A, Weiss J. J Clin Invest. 1996;97:250–257. doi: 10.1172/JCI118399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazzano-Santoro H, Parent J B, Grinna L, Horwitz A, Parsons T, Theofan G. Infect Immunol. 1994;62:1185–1191. [Google Scholar]
- 19.Heumann D, Gallay P, Betz-Corradin S, Barras C, Baumgartner J, Glauser M J. J Infect Dis. 1993;167:1351–1357. doi: 10.1093/infdis/167.6.1351. [DOI] [PubMed] [Google Scholar]
- 20.Wright S D, Jong M T. J Exp Med. 1986;164:1876–1888. doi: 10.1084/jem.164.6.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 22.Jack R S, Grunwald U, Stelter F, Workalemahu G, Schutt C. Eur J Immunol. 1995;25:1436–1441. doi: 10.1002/eji.1830250545. [DOI] [PubMed] [Google Scholar]
- 23.Elsbach P, Weiss J. Infect Agents Dis. 1995;4:102–109. [PubMed] [Google Scholar]
- 24.Katz S S, Chen K, Chen S, Doerfler M E, Elsbach P, Weiss J. Infect Immunol. 1996;64:3592–3600. doi: 10.1128/iai.64.9.3592-3600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen R C. Methods Enzymol. 1986;133:449–493. doi: 10.1016/0076-6879(86)33085-4. [DOI] [PubMed] [Google Scholar]
- 26.Beamer L J, Carroll S F, Eisenberg D. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 27.Elsbach P, Weiss J, Levy O. In: Inflammation: Basic Principles and Clinical Correlates. Gallin J I, Snyderman R, Nathan C, editors. New York: Raven; 1997. [Google Scholar]
- 28.Gray P W, Flaggs G, Leong S R, Gumina R J, Weiss J, Ooi C E, Elsbach P. J Biol Chem. 1989;264:9505–9509. [PubMed] [Google Scholar]
- 29.van Oss C J, Gillman C F. J Reticuloendothel Soc. 1972;12:283–292. [PubMed] [Google Scholar]
- 30.Weiss J, Victor M, Stendahl O, Elsbach P. J Clin Invest. 1982;69:959–970. doi: 10.1172/JCI110535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss J, Kao L, Victor M, Elsbach P. J Clin Invest. 1985;76:206–212. doi: 10.1172/JCI111947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss J, Inada M, Elsbach P, Crowl R M. J Biol Chem. 1994;269:26331–26337. [PubMed] [Google Scholar]