Abstract
The AP-1 transcription factor is a dimeric protein complex formed primarily between Jun (c-Jun, JunB, JunD) and Fos (c-Fos, FosB, Fra-1, Fra-2) family members. These distinct AP-1 complexes are expressed in many cell types and modulate target gene expression implicated in cell proliferation, differentiation, and stress responses. Although the importance of AP-1 has long been recognized, the biochemical characterization of AP-1 remains limited in part due to the difficulty in purifying full-length, reconstituted dimers with active DNA-binding and transcriptional activity. Using a combination of bacterial coexpression and epitope-tagging methods, we successfully purified all 12 heterodimers (3 Jun × 4 Fos) of full-length human AP-1 complexes as well as c-Jun/c-Jun, JunD/JunD, and c-Jun/JunD dimers from bacterial inclusion bodies using one-step nickel-NTA affinity tag purification following denaturation and renaturation of coexpressed AP-1 subunits. Coexpression of two constitutive components in a dimeric AP-1 complex helps stabilize the proteins when compared with individual protein expression in bacteria. Purified dimeric AP-1 complexes are functional in sequence-specific DNA binding, as illustrated by electrophoretic mobility shift assays and DNase I footprinting, and are also active in transcription with in vitro-reconstituted human papillomavirus (HPV) chromatin containing AP-1-binding sites in the native configuration of HPV nucleosomes. The availability of these recombinant full-length human AP-1 complexes has greatly facilitated mechanistic studies of AP-1-regulated gene transcription in many biological systems.
Keywords: AP-1, FLAG tag, hexahistidine tag, inclusion bodies, affinity purification
Introduction
Activator protein-1 (AP-1) is a ubiquitous cellular transcription factor implicated in a wide spectrum of biological processes, including cell proliferation and differentiation, embryonic development, and tumorigenesis [1-3]. In mammalian cells, AP-1 complexes are reconstituted mainly by members of Jun and Fos family proteins, in which each member has a conserved basic region with an adjacent leucine-zipper motif. While the leucine zipper is essential for protein dimerization, the basic region contacts specific DNA sequences often through a consensus, TGA(G/C)TCA, known as the TPA (12-O-tetradecanoylphorbol-13-acetate)-responsive element (TRE). In the Jun family, there are three members: c-Jun, JunB, and JunD. Each of these members can form a homodimer, or heterodimerize with one another within the same family or with members of the Fos family. However, none of the Fos family members (c-Fos, FosB, Fra-1, and Fra-2) form homodimers or heterodimers with the same family members [3]. Many biochemical properties of AP-1 have been elucidated in vitro via the use of truncated proteins containing only the DNA-binding/dimerization domain or by using dimeric partners derived from different species [5-12]. These studies have shed light on the functional role of AP-1 in regulating target gene expression. However, whether the defined DNA-binding and transcriptional activity is truly reflected in the homologous full-length dimeric AP-1 complexes remains to be investigated.
AP-1 is an immediate response gene product regulated by growth factors, cytokines, reactive oxygen species, and various environmental cues [3]. These distinct signaling events modulate AP-1 activity often through posttranslational modifications, including phosphorylation and redox modulation as well as other undefined covalent linkages [13,14]. Since most of purified AP-1 complexes are not readily available, it remains elusive whether many signaling pathways converging on AP-1 in fact directly regulate the DNA-binding activity of AP-1 via covalent modifications. Likewise, many viral systems are also regulated by AP-1. In human papillomavirus (HPV)-infected tissues, propagation of HPV virions is tightly linked to skin differentiation [15]. Interestingly, components of AP-1 subunits are differentially expressed throughout these morphologically distinct skin layers and appear to play an essential role in modulating HPV gene expression and viral assembly [1]. It is intriguing but also puzzling whether these distinct AP-1 complexes are functionally unique or redundant in regulating various cellular and viral promoter activity. To unravel this central issue, we first established an in vitro-reconstituted HPV chromatin transcription system in which transcription from silenced HPV chromatin is dependent on the presence of exogenous c-Jun/c-Fos heterodimers [16]. The availability of this AP-1-dependent chromatin transcription system allows us to define the biochemical properties of distinct AP-1 complexes in a cell-free transcription system that faithfully recapitulates the positioning of HPV chromatin typically seen in vivo [16].
Nevertheless, purification of recombinant full-length human AP-1 complexes remains technically challenging. The preparation of hexahistidine-tagged full-length human c-Fos has not been successful until the introduction of a rare ArgtRNA expression plasmid into E. coli, which confers bacteria the ability to recognize infrequently encountered arginine codons found in human c-Fos [17]. An equal molar ratio of hexahistidine-tagged c-Fos and untagged full-length human c-Jun, each enriched in and quantified from respectively solubilized inclusion bodies, was mixed, denatured, renatured, and then subjected to nickel affinity tag purification. This process, although it has been successful in generating a dimeric c-Jun/c-Fos complex with functional DNA-binding and transcriptional activity [16,17], has also presented several problems. First, exogenously expressed c-Fos tends to be degraded in E. coli. Thus the degradation products, in addition to the full-length protein, are frequently found in purified heterodimers. Second, purified c-Jun/c-Fos may not contain a stoichiometric amount of individual subunits as originally estimated, in part due to the quantification error and also the degradation of c-Fos. To overcome these difficulties, we adapted a coexpression strategy for in vivo reconstitution of distinct AP-1 complexes inside the bacterial cell to facilitate the production of recombinant human AP-1 complexes. The strategy of coexpression has been successful in increasing the stability of coexpressed subunits and in improving the efficiency of multiprotein complex formation [18,19]. We took advantage of the polycistronic expression system because once expression plasmids for individual AP-1 dimers are constructed, we can simply transform a single plasmid into E. coli for protein expression and the in vivo-reconstituted AP-1 complex can be easily purified by affinity tag-based purification protocols. The coexpression system we employed consists of a polycistronic expression vector pST39 and a monocistronic transfer vector pET3aTr, which contains several paired restriction enzyme-cutting sites flanking a translation cassette harboring the desired protein-coding sequence [19]. These paired restriction sites facilitate the cloning of desired AP-1-coding sequences to each assigned cassette that is also flanked by the same restriction sites. Another advantage for the polycistronic expression system is that each of the inserted sequence is preceded by an E. coli ribosome-binding site (RBS) sequence. This design enables T7 RNA polymerase-generated polycistronic transcripts, which contain tandem protein-coding sequences, to be efficiently translated by the E. coli translational machinery. Here, we described for the first time purification of fifteen recombinant full-length human AP-1 dimeric complexes with active DNA-binding and transcriptional activity using bacterial coexpression and affinity tagging methods.
Materials and methods
Plasmid constructions
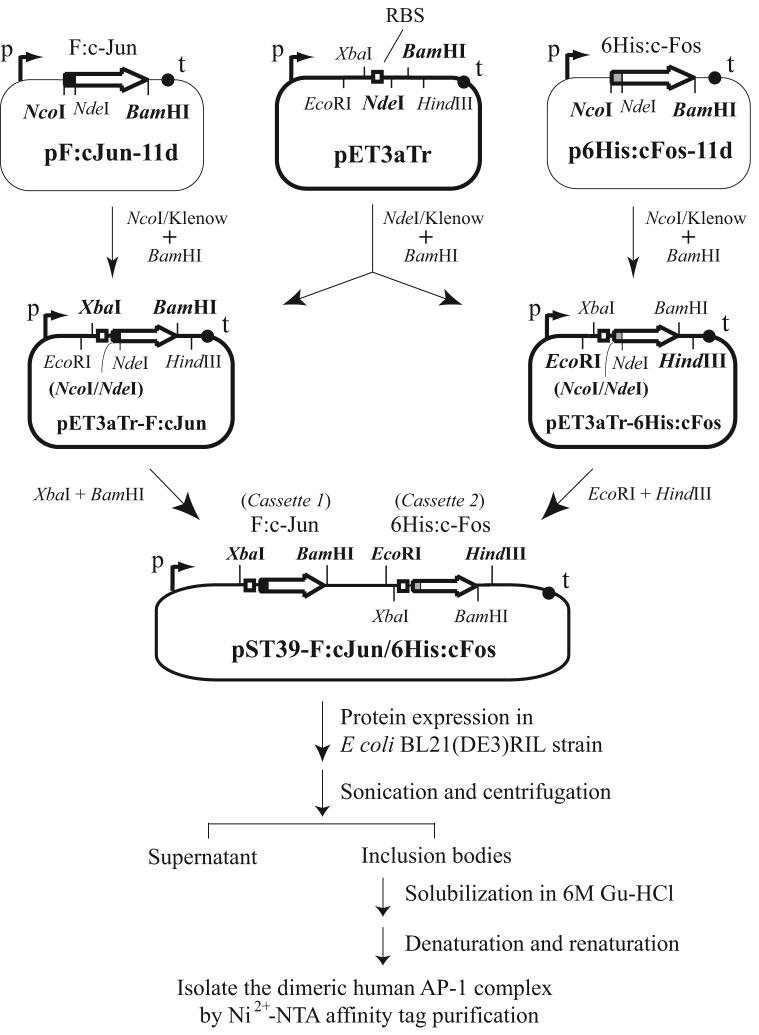
Three stages were involved in the construction of polycistronic bacterial expression plasmids for distinct dimeric human AP-1 complexes (see outline in Figure 1). At the first stage, the coding sequence of human c-Jun, c-Fos, Fra-1, Fra-2, JunB, and JunD was amplified respectively by PCR from pET-Jun and pET-6His-c-Fos [17], pCMV-Fra1 and pCMV-Fra2 [20], pMT3-HA:JunB (a gift from Dr. Yu-Chung Yang at Case Western Reserve University), and pcDNA3.1-hJunD [21], using a primer pair with an NdeI site-containing sense primer annealing to the 5' end and a BamHI site-containing antisense primer hybridizing to the 3' end of individual open reading frame. The DNA fragment carrying the human FosB-coding sequence was amplified from a human brain cDNA library. All the paired primers used for PCR amplification of individual human AP-1 subunits were listed in Table 1. The PCR fragment was then swapped with the TBP insert in pF:TBP-11d [22] or p6His:TBP-11d [23], between NdeI and BamHI sites, to generate an N-terminal FLAG or hexahistidine tag-linked AP-1-coding sequence. In these plasmids, the FLAG or hexahistidine tag sequence was flanked by 5' NcoI and 3' NdeI sites, followed by the inserted AP-1 subunit-coding sequence. The constructs were named pF:cJun-11d, p6His:cJun-11d, pF:cFos-11d, p6His:cFos-11d, and so forth, with F: and 6His: indicating FLAG and hexahistidine tag-linked cDNA sequences, respectively. All the cloned cDNAs were confirmed by DNA sequencing. A point mutation found at the 32nd nucleotide in the original c-Jun-coding region that accidentally changes the encoded amino acid from aspartate (GAC) to glycine (GGC) was corrected by site-directed mutagenesis.
Fig. 1.
Outline of the steps involved in the construction of an AP-1 polycistronic expression plasmid and the procedures for purification of recombinant human AP-1 complexes. The transfer vector pET3aTr contains the T7 promoter (p), terminator (t), ribosome-binding site (RBS) and paired restriction enzyme-cutting sites flanking the coding sequence for FLAG-tagged c-Jun (F:c-Jun) or hexahistidine-tagged c-Fos (6His:c-Fos). The NcoI and NdeI sites in pET3aTrF:cJun and pET3aTr-6His:cFos, destroyed following Klenow filled-in reactions, are indicated by parenthesis. In the purification process, guanidine hydrochloride (Gu-HCl) was used to solubilize bacterial inclusion bodies. For details, see Materials and methods.
Table 1.
Primers for cloning of cDNAs encoding each member of human Jun and Fos family proteins
| Primer name | Sequence |
|---|---|
| h-c-Jun (S-NdeI) | 5'-AAGTCGACATATGACTGCAAAGATGGAA-3' |
| h-c-Jun (AS-BamHI) | 5'-AACTCGAGGATCCTCAAAATGTTTGCAACTG-3' |
| h-JunB (S-NdeI) | 5'-AAGTCGACATATGTGCACTAAAATGGAA-3' |
| h-JunB (AS-BamHI) | 5'-TTCTCGAGGATCCTCAGAAGGCGTGTCCCTT-3' |
| h-JunD (S-NdeI) | 5'-AAGTCGACATATGGAAACACCCTTCTAC-3' |
| h-JunD (AS-BamHI) | 5'-AACTCGAGGATCCTCAGTACGCCGGGACCTG-3' |
| h-c-Fos (S-NdeI) | 5'-AAGTCGACATATGATGTTCTCGGGCTTC-3' |
| h-c-Fos (AS-BamHI) | 5'-AACTCGAGGATCCTCACAGGGCCAGCAGCGTG-3' |
| h-FosB (S-NdeI) | 5'-AAGTCGACATATGTTTCAGGCTTTCCCC-3' |
| h-FosB (AS-BamHI) | 5'-AACTCGAGGATCCTCACCGAGCGAGGAGGGA-3' |
| h-Fra-1 (S-NdeI) | 5'-AAGTCGACATATGTTCCGAGACTTCGGG-3' |
| h-Fra-1 (AS-BamHI) | 5'-AACTCGAGGATCCTCACAAAGCGAGGAGGGT-3' |
| h-Fra-2 (S-NdeI) | 5'-AAGTCGACATATGTACCAGGATTATCCC-3' |
| h-Fra-2 (AS-BamHI) | 5'-TTCTCGAGGATCCTTACAGAGCCAGCAGAGT-3' |
At the second stage, the tagged cDNA insert was subcloned from pET-11d to pET3aTr [19] by cleaving its 5' end with NcoI, blunt-ended with the Klenow enzyme, and then its 3' end by BamHI; the released cDNA was cloned between NdeI/Klenow-BamHI-linearized pET3aTr. The resulting transfer plasmids were accordingly named pET3aTr-F:cJun, pET3aTr-6His:cJun, pET3aTr-F:cFos, pET3aTr-6His:cFos, and etc. At the third stage, a dimeric AP-1 polycistronic expression plasmid was created by cloning from pET3aTr derivatives an FLAG-tagged Jun cDNA (e.g., F:c-Jun) into the first cassette between XbaI and BamHI sites, and a hexahistidine-tagged Fos or Jun family members (e.g., 6His:c-Fos) into the second cassette between EcoRI and HindIII of pST39 [19]. Fifteen human AP-1 polycistronic expression plasmids, generated as described above, were summarized in Table 2.
Table 2.
Polycistronic plasmids expressing dimeric human AP-1 complexes
| Polycistronic plasmid | 1st cassette | 2nd cassette |
|---|---|---|
| pST39-F:cJun/6His:cFos | F:c-Jun | 6His:c-Fos |
| pST39-F:cJun/6His:FosB | F:c-Jun | 6His:FosB |
| pST39-F:cJun/6His:Fra1 | F:c-Jun | 6His:Fra-1 |
| pST39-F:cJun/6His:Fra2 | F:c-Jun | 6His:Fra-2 |
| pST39-F:JunB/6His:cFos | F:JunB | 6His:c-Fos |
| pST39-F:JunB/6His:FosB | F:JunB | 6His:FosB |
| pST39-F:JunB/6His:Fra1 | F:JunB | 6His:Fra-1 |
| pST39-F:JunB/6His:Fra2 | F:JunB | 6His:Fra-2 |
| pST39-F:JunD/6His:cFos | F:JunD | 6His:c-Fos |
| pST39-F:JunD/6His:FosB | F:JunD | 6His:FosB |
| pST39-F:JunD/6His:Fra1 | F:JunD | 6His:Fra-1 |
| pST39-F:JunD/6His:Fra2 | F:JunD | 6His:Fra-2 |
| pST39-F:cJun/6His:cJun | F:c-Jun | 6His:c-Jun |
| pST39-F:cJun/6His:JunD | F:c-Jun | 6His:JunD |
| pST39-F:JunD/6His:JunD | F:JunD | 6His:JunD |
Purification of recombinant dimeric human AP-1 complexes from bacterial inclusion bodies
E. coli BL21(DE3)RIL (Stratagene) harboring individual AP-1 polycistronic expression plasmid (see Table 2) was grown in one liter of TBM9 media (10 g/L of Bacto tryptone, 5 g/L of NaCl, 1 g/L of NH4Cl, 3 g/L of KH2PO4, 6.7 g/L of Na2HPO4.H2O, 4 g/L of glucose, and 1 mM MgSO4) plus 100 μg/mL ampicillin at 37 °C to OD600 around 0.6-0.8 and induced with 0.3 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 15 °C for 3 more hours. Bacterial cells were then pelleted and resuspended in 30 ml of transfer buffer [17] containing 20 mM Tris-HCl (pH 7.9 at room temperature), 20% of glycerol, 1 mM EDTA, 5 mM MgCl2, 0.1 M NaCl, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin, plus 0.1% NP-40. Resuspended cells were sonicated with a Branson Sonifier 450 equipped with a 0.5-inch horn (at output control 8, 30% duty cycle) for 30 bursts, chilled in ice water, and repeated for another four times. Following sonication, the solution was centrifuged at 4 °C, 20,000 rpm for 30 minutes with a Beckman JA-25.50 rotor. The precipitated material, which contains inclusion bodies, was resuspended in 10 ml of transfer buffer (with DTT increased to 5 mM) and dispersed by sonication using a 0.1-inch micro tip (at output control 8, 30% duty cycle) for 30 bursts, ice-chilled in between, and repeated for a total of three times. Inclusion bodies were then pelleted by centrifugation at 4 °C, 15,000 rpm for 10 minutes with a Beckman JA-25.50 rotor. This process (i.e., resuspension, sonication, and centrifugation) for washing the inclusion bodies was repeated for 5 times with the last three washes done by pipetting instead of sonication. The inclusion bodies, after the final wash, were resuspended in 20 ml of buffer A (20 mM Tris-HCl, pH 7.9 at room temperature, 1 mM EDTA, 1 mM DTT, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin) containing 6 M guanidine hydrochloride (Gu-HCl) and dissolved by stirring in a beaker at 4 °C overnight. An aliquot of supernatant, following centrifugation at 4 °C, 15,000 rpm for 10 minutes with a Beckman JA-25.50 rotor, was analyzed by 10% SDS-PAGE and quantified by Coomassie blue staining using bovine serum albumin (BSA) as standards. Solubilized proteins, after adjusting the concentration to ∼200 ng/μl with buffer A plus 6 M guanidine-HCl, was dialyzed sequentially at 4 °C for 3 hours each against 50 volumes of buffer B (20 mM Tris-HCl, pH 7.9 at room temperature, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, and 1 M NaCl) containing 7 M, 1 M and no urea, respectively, and then against buffer C (20 mM Tris-HCl, pH 7.9 at room temperature, 10% glycerol, 0.1 M NaCl, 1 mM DTT, and 0.5 mM PMSF) overnight as described [17]. After centrifugation at 4 °C, 15,000 rpm for 10 minutes with a Beckman JA-25.50 rotor, the supernatant was dispensed into two 15-ml tubes and each incubated with 0.2 ml of Ni2+-NTA agarose beads (Qiagen) at 4 °C for 2 hours. Protein-bound beads were centrifuged at 4 °C, 3,000 rpm for 5 minutes with a Sorvall H-6000A rotor and incubated with 10 ml of buffer C containing 20 mM imidazole by rotation at 4 °C for 5 minutes. The beads were combined and transferred to a microcentrifuge spin column and spun at 4 °C, 10,000 rpm for 1 minute. The dried beads were resuspended in 0.6 ml of buffer C containing 100 mM imidazole by rotation at 4 °C for 30 minutes. Eluted proteins were collected after centrifugation at 4 °C for 1 minute and designated as the first elution. This process was repeated for another two times for the collection of second and third elutions.
The identities of purified AP-1 complexes (i.e., first, second and third elutions) were confirmed by Western blotting with anti-hexahistidine polyclonal antibodies (sc-804, Santa Cruz) for detecting hexahistidine-tagged Fos and Jun family proteins or with anti-FLAG M2 monoclonal antibody (Sigma) for monitoring FLAG-tagged Jun family proteins present in the dimeric AP-1 complex.
Purification of individually expressed human c-Jun and hexahistidine-tagged human c-Fos (6His:c-Fos) and reconstitution of dimeric c-Jun/6His:c-Fos were performed as described previously [17].
Electrophoretic mobility shift assay (EMSA)
The DNA fragment (117 bp) containing an AP-1 binding site TGACTAA [24], derived from the upstream regulatory region (URR) of HPV type 11 (HPV-11), was amplified by PCR using the HPV-11 URR-containing p7072-70GLess/I+ DNA template [25] with a primer pair (sense strand 5'-ACATATTGCCCTGCCAAG-3', and antisense strand 5'-CTTTGGCTGCAATCCACA-3') and end-labeled with [γ-32P]ATP by T4 polynucleotide kinase. Five fmole of the 32P-labeled probe, purified by passing through a MicroSpin G-25 column (GE Healthcare), was used for EMSA as described [26] by incubating with 10 ng of the purified AP-1 complex in a 10-μl reaction containing 10% glycerol, 10 mM HEPES-Na (pH 7.9), 15 mM DTT, 0.2 mM EDTA, 4 mM MgCl2, 0.1 mg/ml BSA, 70 mM NaCl, and 100 ng of poly[d(I-C)] (Roche) at 30 °C for 40 minutes. For oligo competition, 10- or 100-fold excess of unlabelled DNA fragments containing the wild-type or mutated AP-1 site was additionally included at the beginning of the reaction. For antibody-supershift assays, 1 μl containing 68 or 200 ng of rabbit polyclonal antibodies against the hexahistidine tag (sc-804, Santa Cruz) or the N-terminal 79 residues of human c-Jun (sc-1694, Santa Cruz) was added 10 minutes prior to the termination of the reaction. The mixture was then resolved on a 4% nondenaturing polyacrylamide gel, prepared in 5% glycerol-containing 0.25X Tris-borate-EDTA (TBE) buffer, at 80 V room temperature for 2 hours in 0.25X TBE running buffer. The gel was dried and visualized following autoradiography.
DNase I footprinting
The DNA fragment, spanning nucleotides 7688-7921 of the HPV-11 URR that includes previously identified AP-1 binding sites [24], was generated by PCR amplification using p7072-70GLess/I+ with a BamHI site-containing sense primer (5′-GAATTCGGATCCTGCCAAGTATCTTGCCAACA-3′) and a HindIII site-containing antisense primer (5′-TCTAGAAGCTTATATGTAGGGTGTGGGTAAC-3′) and end-labeled with [γ-32P]ATP by T4 polynucleotide kinase. The labeled fragment was then digested with BamHI to prepare a single end-labeled probe, which was further purified by passing through a MicroSpin G-25 column. For DNase I footprinting, 21 fmol of the probe was incubated in a 25-μl reaction [27] containing a different amount (0, 5, 20, and 80 ng) of F:c-Jun/6His:c-Fos or F:c-Jun/6His:FosB at 30 °C for 30 minutes and then digested with 0.01 units of DNase I (Invitrogen) for 2 minutes at room temperature. The reactions products were processed and separated on a 6% polyacrylamide/8% urea gel as described [27] and analyzed by Typhoon 9200 PhosphorImager (GE Healthcare).
In vitro transcription assay
In vitro-reconstituted HPV chromatin was prepared by incubating 1.28 μg of p7072-70GLess/I+ with 3.3 μg of purified HeLa core histones, 3.6 μg of NAP-1 histone chaperone, and 250 ng of ACF chromatin assembly factor at 27 °C for 4 hours according to the published protocol [16]. For the order-of-addition transcription experiment, the assembled chromatin was first preincubated with 30 ng of p300 and 30 μM acetyl-CoA, in the absence or presence of 20 ng of AP-1, at 30 °C for 20 minutes. HeLa nuclear extract (∼80 μg) and pMLΔ53 (10 ng) were then added and incubated at 30 °C for another 20 minutes. Transcription was initiated by the addition of nucleoside triphosphate mix [16] and continued at 30 °C for 60 minutes. Synthesized transcripts were processed and quantified by autoradiography using Typhoon 9200 PhosphorImager as previously described [16].
Results
Generation of polycistronic bacterial expression plasmids for distinct human AP-1 complexes
To purify recombinant full-length human AP-1 complexes, we applied a polycistronic bacterial expression system and epitope tagging for coexpression of the dimeric components in the same bacteria and purified the assembled complexes from inclusion bodies using affinity tag purification as outlined in Figure 1 (for details, see Materials and methods). In this scheme, coexpression of two constitutive subunits in an AP-1 complex, as exemplified by c-Jun/c-Fos, was initiated by generating a bacterial expression plasmid (in the backbone of pET-11d) containing the coding sequence for FLAG-tagged c-Jun (F:c-Jun) and hexahistidine-tagged c-Fos (6His:c-Fos), respectively. The affinity tag-linked cDNA was excised from the resulting construct (i.e., pF:cJun-11d or p6His:cFos-11d) and cloned into the pET3aTr transfer vector to create pET3aTr-F:cJun or pET3aTr-6His:cFos. The coding sequences for F:c-Jun and 6His:cFos were then isolated separately from the transfer vector and cloned into cassette 1 and cassette 2 of the pST39 polycistronic expression plasmid using the designated enzyme pairs as indicated (see Fig. 1). Coexpression of F:c-Jun and 6His:c-Fos was conducted in the bacterial BL21(DE3)RIL strain that harbors specific tRNAs for recognition of human arginine, isoleucine and leucine codons.
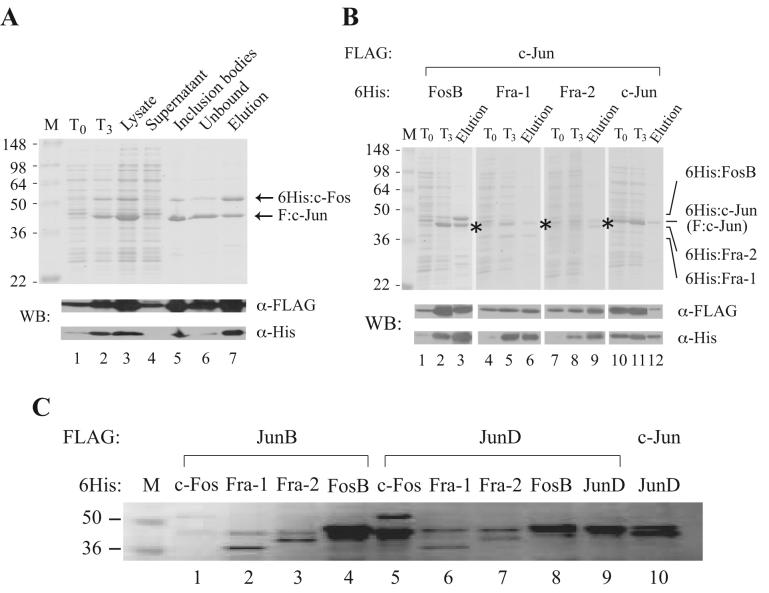
As shown in Figure 2A, expression of F:c-Jun and 6His:c-Fos proteins before (T0) and after IPTG induction for 3 hours (T3) was monitored by Coomassie blue staining and Western blotting with anti-FLAG or anti-hexahistidine tag antibodies. Although leaky expression was detected by Western blotting at T0, a significant induction of F:c-Jun and 6His:c-Fos was visible at T3 even by Coomassie blue staining (lanes 1 vs. 2). The total lysate (lane 3), prepared by sonication, was then separated into supernatant (lane 4) and inclusion bodies (lane 5). Clearly, the majority of overexpressed proteins were found in inclusion bodies with high purity (lanes 4 vs. 5). For purification of F:c-Jun/6His:c-Fos heterodimers, inclusion bodies were solubilized, denatured, renatured, and subjected to Ni2+-NTA binding (see outline in Fig. 1). The dimeric AP-1 complex was finally eluted from the beads in 100 mM imidazole-containing buffer. Interestingly, an equal ratio of F:c-Jun and 6His:c-Fos was present in the purified AP-1 complex, even though F:c-Jun expressed from the first cassette was apparently in excess of 6His:c-Fos expressed in the second cassette (compare lane 7 with lanes 2, 3, 5 and 6), indicating a successful purification of a dimeric AP-1 complex based on the affinity tag introduced into the coding region of the second cassette that tends to have a lower expression efficiency compared to that from the first cassette. The identity of the heterodimeric AP-1 complex was further confirmed by Western blotting with anti-FLAG and anti-hexahistidine antibodies (see Fig. 2A, bottom two strips).
Fig. 2.
Expression and purification of recombinant human AP-1 complexes monitored by Coomassie blue staining and Western blotting. (A) Tracking of F:c-Jun and 6His:c-Fos expression through the purification process. Protein samples were separated by 10% SDS-PAGE and visualized after Coomassie blue staining (top panel) or Western blotting (WB) with anti-FLAG or anti-hexahistidine antibodies (bottom two strips). Lanes 1 and 2 are aliquots of protein samples taken before (T0) and after 3-hour IPTG induction (T3). Lane 3, crude bacterial lysate taken after sonication; lane 4, supernatant taken after centrifugation of the crude lysate; lane 5, sample taken after solubilization of inclusion bodies in 6 M guanidine-HCl solution; lane 6, unbound fraction following Ni2+-NTA binding; lane 7, purified F:c-Jun/6His:c-Fos from the first elution. Protein size markers (in kDa) are indicated on the left. (B) Expression and purification of c-Jun-containing AP-1 complexes. The position of affinity-tagged c-Jun is indicated by asterisk. (C) Coomassie blue-stained gel of purified AP-1 complexes. Purified protein complexes were separated and visualized as described in (A).
The same purification strategy was used to monitor the expression of other c-Jun heterodimers, including c-Jun/FosB, c-Jun/Fra-1 and c-Jun/Fra-2, as well as the c-Jun/c-Jun homodimer. Similar to c-Jun/c-Fos, induction of these dimeric c-Jun-containing complexes was clearly seen after IPTG addition for 3 hours and expression of F:c-Jun from the first cassette is generally higher than the hexahistidine-tagged proteins expressed from the second cassette (Fig. 2B, compare lanes 1 and 2, 4 and 5, 7 and 8, and 10 and 11). Nevertheless, a stoichiometric amount of F:c-Jun and its hexahistidine-tagged partners was found in these purified AP-1 complexes (Fig. 2B, lanes 3, 6, 9 and 12). Ten additional AP-1 complexes, including JunB/c-Fos, JunB/FosB, JunB/Fra-1, JunB/Fra-2, JunD/c-Fos, JunD/FosB, JunD/Fra-1, JunD/Fra-2, JunD/JunD, and c-Jun/JunD, were similarly purified from bacterial inclusion bodies by the same affinity tagging and Ni2+-NTA purification scheme and the resulting complexes were shown in Figure 2C following Coomassie blue staining.
Coexpression stabilizes overexpressed JunB
To examine whether coexpression of dimeric protein partners indeed helps stabilize an easily degradable protein, we expressed F:JunB either individually or in complex with 6His:c-Fos. As shown in Figure 3, very little full-length F:JunB was detected by Western blotting, since most of the F:JunB protein was degraded when expressed from a monocistronic expression plasmid (lanes 1 and 2). In contrast, a significant amount of intact F:JunB was detected when the protein was coexpressed with 6His:c-Fos from a polycistronic expression plasmid (Fig. 3, lanes 3 and 4), indicating the strategy of coexpression could indeed enhance the stability of the labile JunB protein in E. coli.
Fig. 3.
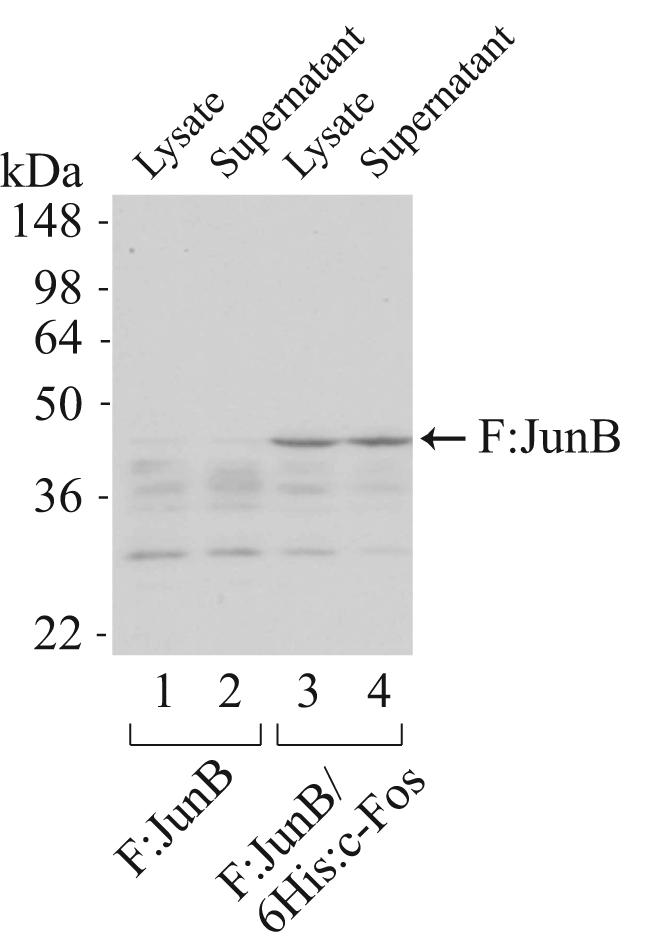
Coexpression enhances the stability of full-length human JunB expressed in E. coli. Bacterial lysate and sonication supernatant taken from individually expressed F:JunB or coexpressed F:JunB/6His:c-Fos were separated by 10% SDS-PAGE and analyzed by Western blotting with anti-FLAG M2 monoclonal antibody. Lanes 1 and 2, F:JunB expressed from a monocistronic plasmid, pF:JunB-11d, which carries the coding sequence of FLAG-tagged human JunB. Lanes 3 and 4, full-length F:JunB stabilized by coexpressed 6His:c-Fos.
Recombinant human AP-1 complexes exhibit sequence-specific DNA-binding activity
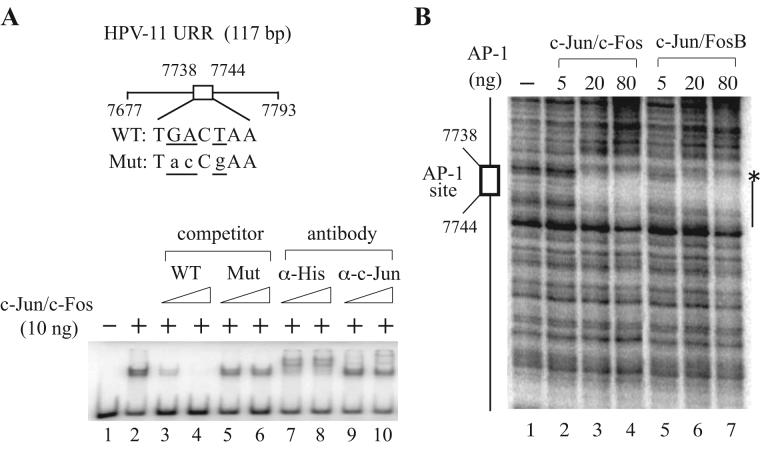
To define whether recombinant AP-1 complexes purified from bacterial inclusion bodies possess DNA-binding activity following denaturation and renaturation, we performed electrophoretic mobility shift assays (EMSA) using purified F:c-Jun/6His:c-Fos with a DNA probe containing a well-characterized AP-1-binding site (TGACTAA) derived from HPV-11 nucleotides 7738-7744 [24]. As shown in Figure 4, a specific AP-1-bound protein-DNA complex was competed away by increasing amounts of non-radioactive cold competitors harboring the wild-type but not mutated AP-1-binding site (lanes 1-6). The presence of dimeric F:c-Jun/6His:c-Fos was illustrated by antibody-supershift experiments using anti-hexahistidine or anti-c-Jun antibodies, which induced formation of higher molecular DNA-binding species (Fig. 4, lanes 7-10). An inefficient supershift observed with anti-c-Jun antibodies, raised against amino acids 1-79 of human c-Jun, suggests that the N-terminal region of c-Jun is buried in the dimeric c-Jun/c-Fos complex. This is consistent with our inability to supershift the protein-DNA complex using anti-FLAG M2 monoclonal antibody (data not shown). Undoubtedly, the N-terminus of c-Fos is likely exposed on the surface of the c-Jun/c-Fos complex, since the AP-1-bound probe was completely up-shifted by anti-hexahistidine antibodies.
Fig. 4.
Purified AP-1 complexes possess sequence-specific DNA-binding activity. (A) Electrophoretic mobility shift assays (EMSA) performed with F:c-Jun/6His:c-Fos. EMSA was conducted as described in Materials and methods using a radiolabeled AP-1 site-containing DNA fragment (117 bp) derived from the HPV-11 upstream regulatory region (URR), in the absence (−) or presence (+) of 10 ng of F:c-Jun/6His:c-Fos with or without an increasing amount of wild-type (WT) or mutant (Mut) cold competitors (10- and 100-fold in excess) or anti-hexahistidine or anti-c-Jun antibodies. (B) DNase I footprinting performed with F:c-Jun/6His:c-Fos and F:c-Jun/6His:FosB. DNase I footprinting was carried out with different amounts (0, 5, 20 and 80 ng) of F:c-Jun/6His:c-Fos or F:c-Jun/6His:FosB as described in Materials and methods using a labeled DNA probe containing a well-characterized AP-1 site (indicated by a box) found in the HPV-11 URR. A hypersensitive site (*) and the protected region are depicted on the right.
The sequence-specific DNA-binding activity of purified AP-1 complexes was further demonstrated by DNase I footprinting assays using recombinant F:c-Jun/6His:c-Fos and F:c-Jun/6His:FosB. A clear protection surrounding the same AP-1-binding site explored in EMSA was observed upon the addition of increasing amounts of either dimeric complex (Fig. 4C), indicating that recombinant AP-1 complexes are able to recognize their cognate sequences in a dose-dependent manner.
Recombinant AP-1 complexes are able to activate transcription from HPV chromatin
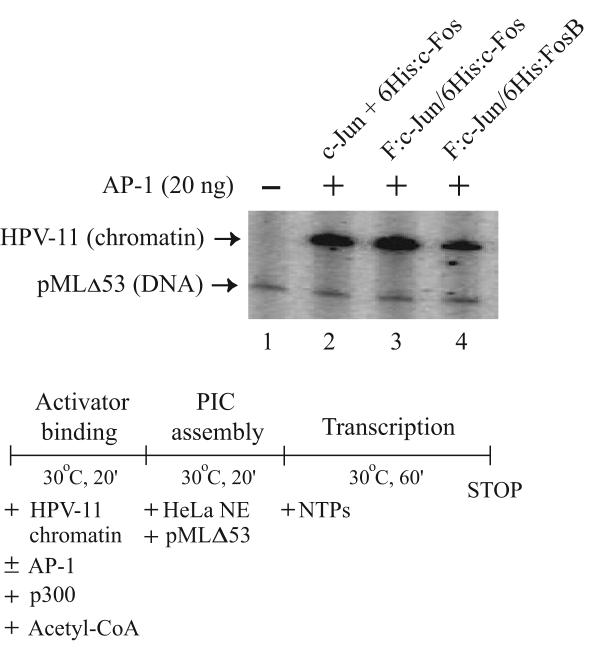
To analyze the transcriptional activity of purified recombinant AP-1 complexes, we conducted a cell-free transcription assay using in vitro-reconstituted HPV chromatin that faithfully recapitulates the positioning of nucleosomes typically observed in vivo [16]. The transcription experiment was carried out in three steps [16,28] to facilitate activator binding and p300-mediated acetylation of chromatin, before the inclusion of transcriptional components provided by HeLa nuclear extract and prior to the initiation of transcription by adding ribonucleoside triphosphates (see outline in Fig. 5). A nucleosome-free DNA template (pMLΔ53) that generates a shorter transcript independent of AP-1 was also included as an internal control. As shown in Figure 5, no transcript could be detected from silenced HPV chromatin, when AP-1 was not included in the reaction (lane 1). However, the transcript resulted from the internal control was synthesized, indicating the transcription machinery was fully active. Interestingly, F:c-Jun/6His:c-Fos purified from the coexpression system (lane 3) had comparable transcriptional activity compared with individually expressed and reconstituted c-Jun and 6His:cFos (lane 2) used in our previous chromatin-dependent transcription study [16]. Recombinant F:c-Jun/6His:FosB could also activate HPV chromatin transcription, although its activity seems to be weaker than that derived from the c-Jun/c-Fos complexes (Fig. 5, lane 4 vs. lanes 2 and 3).
Fig. 5.
Purified AP-1 complexes exhibit transactivation activity on reconstituted HPV chromatin. In vitro-reconstituted HPV-11 chromatin was incubated with individually expressed and reconstituted c-Jun/6His:c-Fos (lane 2), or coexpressed F:c-Jun/6His:c-Fos (lane 3) or F:c-Jun/6His:FosB (lane 4) and p300 and acetyl-CoA at 30°C for 20 minutes. HeLa nuclear extract (NE) and the pMLΔ53 internal DNA control were then added for preinitiation complex (PIC) assembly at 30°C for 20 minutes. Transcription was initiated by the addition of ribonucleoside triphosphates (NTPs) and continued for an hour. Transcripts synthesized from HPV-11 chromatin and pMLΔ53 DNA were analyzed as described in Materials and methods and visualized following autoradiography.
Discussion
Using bacterial coexpression and affinity tag methods, we have successfully purified 15 recombinant full-length human AP-1 complexes with active DNA-binding and transcriptional activity. Purification of these dimeric human AP-1 complexes is facilitated by the construction of bacterial polycistronic expression plasmids carrying pairwise AP-1 subunit-coding sequences in various combinations (see Table 2). The introduction of different affinity tags [29], e.g., FLAG epitope and hexahistidine sequences, also allows protein tracking of individual subunits throughout the entire induction and purification process (see Fig. 2A). Whenever necessary, a second step of affinity purification can be employed to further purify the dimeric complex following hexahistidine tag purification. Nevertheless, in the case of AP-1, the FLAG tag sequence introduced at the N terminus of c-Jun appears to be buried in the dimeric complex (see Fig. 4A), making it unfeasible to conduct sequential affinity tag purification based on the FLAG epitope sequence. This unexpected finding did not thwart our efforts in purifying distinct human AP-1 complexes for the following reasons:
1. Both subunits of human AP-1 complexes were significantly enriched in inclusion bodies with high purity (see Fig. 2A) and without undergoing serious degradation as often seen in the soluble fraction with individually expressed proteins (see Fig. 3).
2. The level of protein expression from the second cassette containing the hexahistidine tag was generally lower than that expressed from the first cassette with the FLAG epitope sequence in most of human AP-1 polycistronic expression plasmids (see Fig. 2A and 2B). This design has made it possible to purify AP-1 complexes with a stoichiometric ratio of dimeric protein partners simply by one-step affinity purification based on the hexahistidine tag sequence introduced at the second cassette. It further minimizes potential contamination by c-Jun homodimers, since the FLAG-tagged Jun family member expressed from the first cassette in an excess amount would not bind to Ni2+-NTA beads.
Nevertheless, it should be mentioned that the stoichiometric ratio of the dimeric components in distinct AP-1 complexes might not always be equal following one-step affinity purification (see Fig. 2C). In that case, additional biochemical and biophysical methods, such as conventional column chromatography, analytic ultracentrifugation, isothermal calorimetry, and differential scanning calorimetry, can be employed to further purify and characterize the complexes to ensure that appropriate hetero- or homodimers are indeed present in the final product.
Purification of recombinant proteins from bacterial inclusion bodies has been widely used for production of many pharmaceutically important proteins [29]. Inclusion bodies provide insulation for overexpressed proteins and protect them from degradation by bacterial proteases, and thus are a rich source for full-length intact proteins. The primary steps involved in the purification of dimeric human AP-1 complexes include construction of AP-1 polycistronic expression plasmids, coexpression of recombinant protein partners in BL21(DE3)RIL, isolation and solubilization of inclusion bodies, denaturation, renaturation, and nickel affinity tag purification (see Fig. 1). This procedure makes it feasible for us to purify various combinations of recombinant full-length human AP-1 complexes that have been very difficult to achieve in the past. These dimeric AP-1 complexes can now be used to define the mechanism of AP-1-mediated target gene regulation and also decipher the role of posttranslational modification in regulating DNA-binding and transcriptional activity of AP-1 as well as its interaction with other cellular protein factors and cofactors implicated in diverse signaling pathways.
Acknowledgments
We thank Paul Dobner for providing pCMV-Fra1 and pCMV-Fra2 plasmids, James Goodrich for pET-Jun and pET-6His-c-Fos, Curt Pfarr for pcDNA3.1-hJunD, Song Tan for pET3aTr and pST39, and Yu-Chung Yang for pMT3-HA:JunB. We are also grateful to Shwu-Yuan Wu for many helpful discussions in the development of the purification protocol. This work is supported in part by grants CA103867 and CA124760 from the National Institutes of Health and is Report CSCN # 033 from University of Texas Southwestern Medical Center Simmons Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckert RL, Crish JF, Banks EB, Welter JF. The epidermis: genes on - genes off. J. Invest. Dermatol. 1997;109:501–509. doi: 10.1111/1523-1747.ep12336477. [DOI] [PubMed] [Google Scholar]
- 2.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 3.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 4.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 5.Gentz R, Rauscher FJ, Abate C, Curran T. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science. 1989;243:1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- 6.Turner R, Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989;243:1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- 7.Abate C, Luk D, Gagne E, Roeder RG, Curran T. Fos and jun cooperate in transcriptional regulation via heterologous activation domains. Mol. Cell. Biol. 1990;10:5532–5535. doi: 10.1128/mcb.10.10.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abate C, Luk D, Gentz R, Rauscher FJ, III, Curran T. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1032–1036. doi: 10.1073/pnas.87.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abate C, Luk D, Curran T. Transcriptional regulation by Fos and Jun in vitro: interaction among multiple activator and regulatory domains. Mol. Cell. Biol. 1991;11:3624–3632. doi: 10.1128/mcb.11.7.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 11.Kerpolla TK, Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland JA, Cook A, Bannister AJ, Kouzarides T. Conserved motifs in Fos and Jun define a new class of activation domain. Genes Dev. 1992;6:1810–1819. doi: 10.1101/gad.6.9.1810. [DOI] [PubMed] [Google Scholar]
- 13.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 14.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehrmann F, Laimins LA. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene. 2003;22:5201–5207. doi: 10.1038/sj.onc.1206554. [DOI] [PubMed] [Google Scholar]
- 16.Wu S-Y, Lee A-Y, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang C-M. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson HA, Goodrich JA. Expression and purification of recombinant human c-Fos/c-Jun that is highly active in DNA binding and transcriptional activation in vitro. Nucleic Acids Res. 2001;29:E98. doi: 10.1093/nar/29.20.e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Schwabe JW, Banayo E, Evans RM. Coexpression of nuclear receptor partners increases their solubility and biological activities. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2278–2283. doi: 10.1073/pnas.94.6.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan S. A modular polycistronic expression system for overepressing protein complexes in Escherichia coli. Protein Expr. Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 20.Harrison RJ, McNeil GP, Dobner PR. Synergistic activation of neurotensin/neuromedin N gene expression by c-Jun and glucocorticoids: novel effects of Fos family proteins. Mol. Endocrinol. 1995;9:981–993. doi: 10.1210/mend.9.8.7476995. [DOI] [PubMed] [Google Scholar]
- 21.Short JD, Pfarr CM. Translational regulation of the JunD messenger RNA. J. Biol. Chem. 2002;277:32697–32705. doi: 10.1074/jbc.M204553200. [DOI] [PubMed] [Google Scholar]
- 22.Chiang C-M, Roeder RG. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Pept. Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 23.Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder RG. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou W, Chow LT, Broker TR. Transcription activities of human papillomavirus type 11 E6 promoter-proximal elements in raft and submerged cultures of foreskin keratinocytes. J. Virol. 1997;71:8832–8840. doi: 10.1128/jvi.71.11.8832-8840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou SY, Wu S-Y, Zhou T, Thomas MC, Chiang C-M. Alleviation of human papillomavirus E2-mediated transcriptional repression via formation of a TATA binding protein (or TFIID)-TFIIB-RNA polymerase II-TFIIF preinitiation complex. Mol. Cell. Biol. 2000;20:113–125. doi: 10.1128/mcb.20.1.113-125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou SY, Wu S-Y, Chiang C-M. Transcriptional activity among high and low risk human papillomavirus E2 proteins correlates with E2 DNA binding. J. Biol. Chem. 2002;277:45619–45629. doi: 10.1074/jbc.M206829200. [DOI] [PubMed] [Google Scholar]
- 27.Chiang C-M, Ge H, Wang Z, Hofmann A, Roeder RG. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerase II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas MC, Chiang C-M. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell. 2005;17:251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 2005;99:303–310. doi: 10.1263/jbb.99.303. [DOI] [PubMed] [Google Scholar]