Abstract
Staphylococcal α-toxin is a 293-residue, single-chain polypeptide that spontaneously assembles into a heptameric pore in target cell membranes. To identify the pore-forming domain, substitution mutants have been produced in which single cysteine residues were introduced throughout the toxin molecule. By attaching the environmentally sensitive dye acrylodan to the sulfhydryl groups, the environment of individual amino acid side chains could be probed. In liposomes, a single 23-amino acid sequence (residues 118–140) was found to move from a polar to a nonpolar environment, indicating that this sequence forms the walls of the pore. However, periodicity in side chain environmental polarity could not be detected in the liposomal system. In the present study, the fluorimetric analyses were extended to physiological target cells. With susceptible cells such as rabbit erythrocytes and human lymphocytes, the 23 central amino acids 118–140 were again found to insert into the membrane; in contrast to the previous study with liposomes, the expected periodicity was now detected. Thus, every other residue in the sequence 126–140 entered a nonpolar environment in a striking display of an amphipathic transmembrane β-barrel. In contrast, human granulocytes were found to bind α-toxin to a similar extent as lymphocytes, but the heptamers forming on these cells failed to insert their pore-forming domain into the membrane. As a consequence, nonfunctional heptamers assembled and the cells remained viable. The data resolve the molecular organization of a pore-forming toxin domain in living cells and reveal that resistant cells can prevent insertion of the functional domain into the bilayer.
Keywords: pore-forming toxin, cysteine-scanning mutagenesis, membrane permeabilization
Formation of transmembrane pores is a major mechanism by which exogenous proteins damage target cells. Membrane permeabilization underlies the action of many bacterioicins (1), larval insecticidal toxins (2), microbial exotoxins (3), and mammalian defense effector systems (4). Pore formation also appears to be required for translocation of several intracellularly acting toxins across lipid bilayers (5).
For α-toxin, a 293-residue single-chain polypeptide that is a major virulence determinant of Staphylococcus aureus (6), the following model for pore assembly accommodating four organizational and conformational states has emerged (6, 7). The first state is the water-soluble, native monomer. The second is the membrane-bound monomer. The third is the heptameric pre-pore, where the pore-forming domain is on the verge of membrane insertion. The final pore configuration results after the functional domain enters the membrane, forming the walls of a hydrophilic channel.
Substitution mutants were previously produced in which single cysteine residues were introduced throughout the toxin molecule (8). By attaching the polarity-sensitive dye acrylodan (9) to the sulfhydryl groups, the environment of individual amino acid side chains in toxin monomers and in membrane-bound oligomers were probed. In liposomes, a single 23-amino acid sequence (residues 118–140) was observed to move from a polar to a nonpolar environment (8), and lipid insertion of this amino acid sequence correlated with pore formation (10). Fluorescence energy transfer data indicated that residue 130 was in immediate proximity with the cytoplasmic membrane face (11). Other structure–function studies yielded data consistent with the model of a cylindrical toxin pore formed by this central molecular domain (12–15).
Heptameric α-toxin oligomers assemble spontaneously in detergent solution (16, 17), and their three-dimensional structure has recently been resolved (18). On the assumption that such oligomers are structurally identical to those formed in lipid bilayers, these data show that the seven amphipathic central domains encompassing amino acid residues 118–140 indeed form the walls of a water-filled channel in the form of a β-barrel (18).
An essential complementary approach to the above studies is the investigation of the molecular conformation of α-toxin heptamers in membranes of physiological cell targets, and the present investigation was undertaken to fill this information gap. In the course of our experiments, it was found that human granulocytes display a natural resistance against the pore-forming action of α-toxin that is not due to hindrance of toxin binding or heptamer assembly. Data from the fluorimetric analyses are presented that demonstrate a striking difference in the orientation of the pore-forming sequence in heptamers assembled on toxin-susceptible versus toxin-resistant cells; this difference correlated with and accounted for the cellular phenotypes.
MATERIALS AND METHODS
Toxin Substitution Mutants.
The production of 44 cysteine substitution mutants has been described (8, 15, 19). Eight additional mutants were produced, three at the N terminus (19) and five completing the sequence 126–140 by PCR mutagenesis according to published procedures (8, 20). The latter mutants were as follows: I132C, G133C, L135C, I136C, and N139C. All acrylodan-labeled mutants except N139C retained full specific activity. Acrylodan-labeled N139C exhibited an approximately 25% reduction in hemolytic activity, but membrane-bound N139C assembled into heptamers to the same extent as other toxin derivatives.
Cells.
Human T-lymphocytes and neutrophil granulocytes were prepared as described (21, 22). A total of 5 × 106 cells/ml in PBS were incubated with α-toxin. Phagocytosis tests and flow cytometry were performed as described (22).
Measurement of K+ Efflux and Intracellular ATP.
For K+ efflux measurements, 107 cells/ml were incubated with α-toxin at 37°C for 60 min. Samples were withdrawn and centrifuged for 5 min at 250 × g, followed by 5 min at 13,000 × g. Potassium concentrations were determined by an atomic absorption spectrophotometer (type AA-5; Varian-Techtron, Melbourne, Australia).
For measurements of cellular ATP, 105 cells in 200 μl of medium were treated with α-toxin (60 min, 37°C) and then lysed with 10 μl of 10% Triton X-100 (without centrifugation). Each lysate was transferred into a vial containing 155 μl of distilled water and 25 μl of luciferase (Boehringer Mannheim), and chemiluminescence was measured in a luminometer (Bioluminat LB 9500; Berthold, Wildbad, Germany). ATP contents of toxin-treated cells were expressed as percent luminescence relative to that of untreated cells (23).
Spectrofluorimetric Studies.
Acrylodan-labeled toxins were incubated with liposomes as described (8). Rabbit erythrocytes (5 × 108 cells/ml) were lysed with 5 μg/ml of labeled toxin. T-lymphocytes or granulocytes (5 × 106 cells/ml) were treated with 20 μg/ml of labeled toxin for 45 min at 37°C. Erythrocyte membranes were pelleted and washed in a bench top microfuge (10,000 × g for 5 min). Lymphocytes and granulocytes were sedimented (2,000 × g for 10 min), washed twice, and resuspended in PBS. Fluorescence emission spectra were recorded in a Spex Fluoromax fluorimeter (wavelength: excitation 365 nm, emission 400–600 nm; scanning interval 1 nm; band passes: excitation 4 nm, emission 2 nm). For all samples, the spectra of appropriate blanks (buffers, liposome, or cell suspension) were recorded and subtracted from the sample spectra (8). A narrow bore cuvette was used to minimize spectral distortions due to light scattering.
RESULTS
Human Granulocytes Display a Primary Resistance Toward the Pore-Forming Action of α-Toxin.
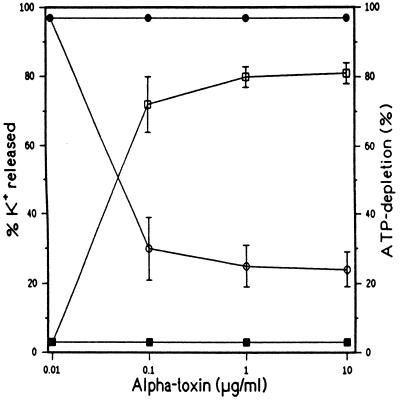
In contrast to T-lymphocytes, human granulocytes can be exposed to high concentrations of α-toxin without becoming permeabilized. No depletion of cellular K+ or ATP (Fig. 1) or inhibition of phagocytic function (data not shown) was observed after incubation of granulocytes with 20 μg/ml α-toxin for 60 min. The primary resistance of human granulocytes toward α-toxin might be due to several different reasons. First, granulocytes might lack toxin-binding sites. Second, oligomerization of bound monomers might be inhibited. Third, heptamers might form that are rapidly moved by endocytosis or shedding. Or fourth, nonfunctional heptamers might assemble on these membranes. The following experiments excluded the first three possibilities.
Figure 1.
Measurement of cellular ATP-depletion and K+-efflux in α-toxin-treated cells. Lymphocytes (open symbols) or granulocytes (filled symbols) were treated with α-toxin for 60 min at 37 C. K+ concentrations (squares) in supernatants were determined. Cells were lysed with Triton X-100 and ATP concentrations in the lysates (circles) were determined. Data represent means ± SD (error bars) for three independent experiments.
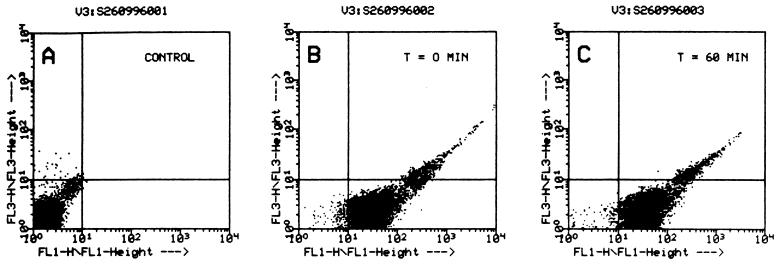
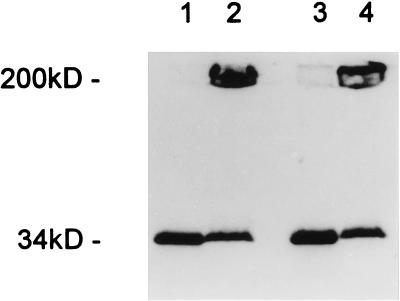
Binding studies were performed using a radioiodinated tracer, and it was found that binding of α-toxin occurred rapidly (essentially complete within 1 min at 25°C) and to a similar extent to both lymphocytes and granulocytes (data not shown). In a second set of experiments, a cysteine-substitution mutant S69C was employed. This toxin derivative could be labeled with either fluorescein-maleimide or biotin-maleimide without loss of pore-forming activity (24). When the fluorescein-labeled derivative was added to lymphocytes or granulocytes, similar fluorescence intensities of the two cell types were observed by flow cytometry (data not shown), indicating similar binding of the toxin at the single-cell level. The S69C mutant was then labeled with biotin-maleimide because this enabled quantitative assessment of its cell surface localization to be performed. Biotinylated S69C toxin was bound to cells, and the surface accessibility of biotin was subsequently probed by incubation of the cells with streptavidin-fluorescein. α-Toxin could be detected on the cell surface over the entire duration of the experiment (60 min) (Fig. 2). Use of labeled S69C-toxin also enabled demonstration that no truncation of the toxin within the heptamers had occurred. Thus, SDS-stable oligomers were present on both lymphocytes and granulocytes, and boiling generated a monomeric 34-kDa α-toxin (Fig. 3). This finding was relevant because it rendered proteolytic nicking as a mechanism for heptamer inactivation improbable. Therefore, lack of permeabilization in granulocytes was due neither to prevention of heptamer assembly nor to proteolytic destruction of the toxin pore.
Figure 2.
Nonfunctional α-toxin molecules bound to granulocytes remain surface-exposed. Granulocytes (5 × 106 cells/ml) in PBS were incubated with 20 μg/ml of biotinylated S69C α-toxin (23) for 5 min at 37°C. Cells were then washed twice, suspended in buffer, and either incubated immediately (B) or after 60 min (C) with streptavidin-fluorescein (5 μg/ml) for 5 min at 37°C, and subjected to flow-cytometric analyses. Control cells (A) incubated with unbiotinylated S69C α-toxin exhibited no fluorescence, whereas cells laden with biotinylated S69C α-toxin assumed a green fluorescence that remained essentially unaltered after they had been incubated in buffer for 60 min (C). Horizontal axis: F1, relative green fluorescence, vertical axis: F3, relative red fluorescence. Propidium iodide was added at 4 μg/ml prior to flow cytometry, and lack of red fluorescence indicated that cells remained impermeable to propidium iodide throughout the experiment.
Figure 3.
SDS/PAGE (UV-transilluminated) demonstrates that α-toxin heptamers form on human lymphocytes (lanes 1 and 2) and granulocytes (lanes 3 and 4). Cells were treated with 20 μg/ml S69C α-toxin labeled with fluorescein-maleimide (23) for 30 min at 37°C, washed, and solubilized in SDS at 95°C to dissociate heptamers (lanes 1 and 3) or solubilized in SDS at room temperature to preserve heptamers (lanes 2 and 4).
Molecular Architecture of α-Toxin Pore in Sensitive Cells.
The emission spectrum of acrylodan attached to single cysteine residues distributed throughout the toxin molecule was analyzed in the monomeric toxin and in heptamers formed on liposomes and cell membranes. A “blue” shift in emission spectrum of the molecule following membrane-binding indicated movement of acrylodan either into a hydrophobic protein pocket or into the lipid environment (8).
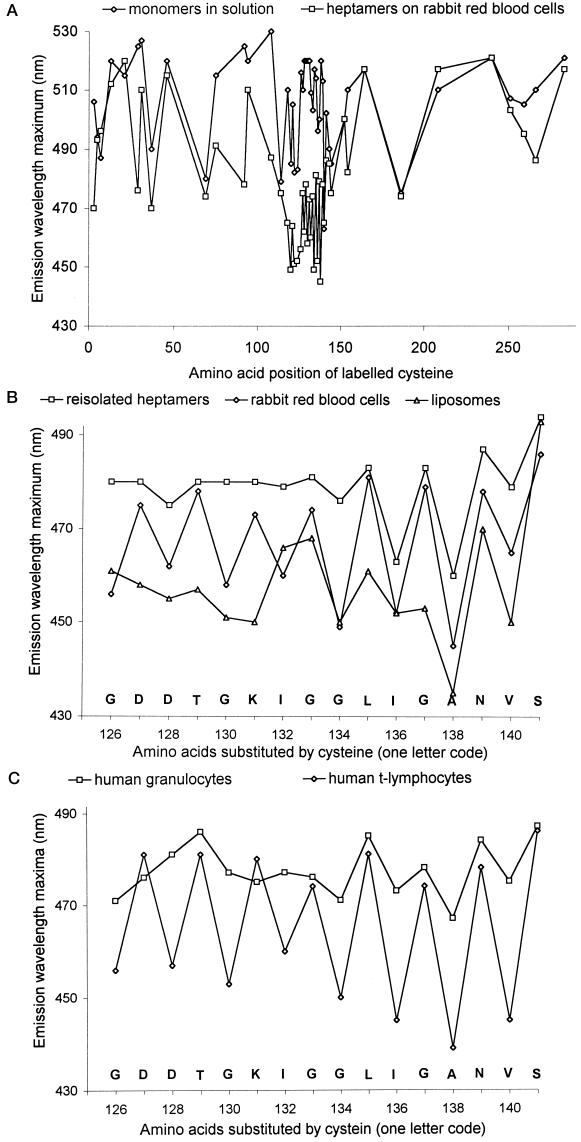
Fluorimetric data were obtained with 49 functionally intact, acrylodan-labeled toxin mutants on erythrocyte membranes. Fig. 4A shows the emission maxima for each derivative in solution and in the membrane-bound heptamer. Acrylodan attached to amino acid residues 118–140 displayed a marked blue shift upon heptamer assembly. The fluorescence patterns resembled those observed in liposomes (8) and were complemented by data from mutants completing the series spanning residues 126–140. There was one important difference: in liposomes, no periodicity in side-chain environmental polarity was discerned along the pore-forming sequence (8). With erythrocyte membranes, the anticipated periodicity was found. With liposomes and erythrocytes, similar blue shifts of all even-numbered residues were observed that were partially reversed upon delipidation with deoxycholate. Previous experimental data (8) indicated that this shift reversal reflected contact of the labeled side chain with lipid in the bilayer. The degree of fluorescent change varied depending on the detergent used, due probably to detergent–lipid exchange. With the odd residues, however, the emission maxima measured on cells were always less shifted than on liposomes, approximating the emission maxima of re-isolated heptamers (Fig. 4B). Partial reversal of the blue shift was also observed with G122C and V124C when heptamers were formed in and re-isolated from erythrocytes; in the same system, shift reversal was not observed with N121C. We were unable to produce mutants N123C and T125C. From these data, a model for the organization of α-toxin on susceptible cells emerged in which only the even residues in the pore-forming domain are in contact with membrane lipids, whereas side chains of the odd residues face toward the pore lumen.
Figure 4.
(A) Fluorescence emission maxima of acrylodan-labeled α-toxin cysteine mutants in solution (⋄) and after heptamer assembly on rabbit erythrocytes (□). An emission blue shift indicates movement of the fluorescence label to a more hydrophobic environment. Significant blue shifts were observed exclusively in the central region spanning residues 118–140. (B) Emission maxima of α-toxin cysteine mutants labeled with acrylodan in the central pore-forming domain. Emission spectra were recorded on liposomes (▵), rabbit erythrocytes (⋄), and heptamers re-isolated in deoxycholate (□). (C) Emission maxima of acrylodan-labeled α-toxin cysteine mutants measured on human granulocytes (□) and human T-lymphocytes (⋄), showing lack of lipid contact of the pore-forming domain in the toxin-resistant granulocytes.
Heptamers Formed on Resistant Cells Fail to Insert the Pore-Forming Sequence into the Bilayer.
The spectrofluorimetric findings were very similar whether rabbit erythrocytes, human lymphocytes, monocytes, or fibroblasts were employed. With human granulocytes, however, a striking difference was discovered exclusively within the pore-forming sequence. Here, the blue shift in acrylodan fluorescence of those residues exposed to the lipid environment on the other cell membranes was absent (Fig. 4C). Apart from this difference, all the other fluorimetric measurements yielded the same results as found with sensitive cells. Thus, these data revealed that assembly of toxin oligomers had occurred up to the final pre-pore stage (see refs. 3 and 7 for schematic depictions), but granulocytes can thwart the lethal membrane insertion step. As a consequence, nonfunctional heptamers form on these cells.
DISCUSSION
The present data demonstrate how fluorescently labeled cysteine substitution mutants of a pore-forming toxin can be utilized to obtain detailed information on the molecular architecture of a membrane-spanning protein domain in physiological cell targets. The rationale is straightforward: a blue shift in fluorescence emission spectrum indicates movement of a given amino acid side chain to a nonpolar environment. If the blue shift is not reversed in delipidated oligomers, this indicates burial of the reporter molecule within a protein pocket. If, however, a partial shift reversal occurs upon delipidation, this indicates immediate contact of the side chain with lipid (8). It is notable that the degree of shift reversal is dependent on the detergent used to replace the lipids (8), which is in accord with the interpretation. The present investigation has generated a number of generally relevant insights. The observation that employment of viable cells is feasible merits attention. Not only does this imply that it is possible to analyze the conformation of the functional domain in true biological targets, but it will now become possible to follow the fate of the fluorescent reporter molecule in cells over extended time periods. This result will be of interest, for example, for the study of repair processes that sometimes occur (25).
A major finding related to the difference in fluorimetric measurements made exclusively within the pore-forming sequence on cells versus liposomes. In the latter, no periodicity in environmental polarity could be detected, and this had raised concerns about the reliability and resolution of the method. In cell membranes, however, the anticipated periodicity was found, and a model thus emerged in which every even residue in the sequence 118–140 contacted lipid, whereas every uneven residue did not. By exclusion, the latter must face the lumen of the pore. Of note, the emission spectra of all labeled residues facing the lumen were blue-shifted relative to the monomers in solution. This finding probably reflected protein–protein interactions within the channel because they remained unchanged after detergent solubilization. The lack of periodicity in side-chain environment previously observed in artificial lipid bilayers could indicate that the β-strands enjoy greater rotational mobility in the membranes of small liposomes that were employed in that study (8). It is indeed known that the vesicle bilayer in small liposomes is strained, and packing of the lipid molecules deviates from that given in large vesicles (26, 27). Repetition of the experiments using large unilamellar liposomes, prepared according to ref. 28, has in fact recently generated data similar to those acquired with the susceptible cells (data not shown).
Comparison of this model with the crystal structure of the heptamer formed in detergent solution reveals superb congruence (18). In the latter, residue 118 represents the starting point from which the next 23 residues would enter the hydrophobic membrane interior, forming an amphipathic β-barrel, with side-chains of even residues in contact with membrane lipids, those of uneven residues facing the pore lumen, and residue 129 forming the turning point at the cytoplasmic membrane face. From previous data, we had indeed concluded that residue 121 projects into the lumen of the channel and that residue 130 resides near the cytoplasmic membrane face (8, 11). Surely, this congruence cannot be a mere coincidence, and we conclude that the three-dimensional structure of the detergent-assembled heptamer most likely is identical to the pore formed on susceptible cells. On resistant cells such as human granulocytes, however, the structure must clearly be different. The present data revealing lack of membrane insertion of the pore-forming domain show the molecular mechanism of cellular resistance to a pore-forming toxin and highlight the usefulness of the fluorimetric approach for the study of molecular toxin organization in cell membranes. The method compares favorably with electron spin resonance measurements using cysteine substitution mutants which have recently permitted the transmembrane orientation of the putative pore-forming α-helix of diphtheria toxin B subunit to be mapped in liposomes (29), and the fluorimetric approach is technically certainly less demanding. The reasons underlying lack of insertion of the stem domain in granulocytes remains to be elucidated. Ongoing experiments indicate that lack of insertion correlates with nonfunctional heptameric pre-pore assembly in several other cases (unpublished data).
Inhibition of membrane insertion may be a mechanism underlying resistance of cells toward other pore-forming proteins. Streptolysin-O occasionally can assemble into nonfunctional polymers (30), and homologous restriction factors inhibit complement (31), in part also by causing formation of nonlytic terminal complexes (32). Moreover, membrane lesions can sometimes be repaired (25, 31). Environmental probing with fluorescent cysteine-substitution mutants is a potentially useful tool which may yield clues on the responsible mechanisms.
Acknowledgments
We thank Susanne Strauch and Brunhilde Moewes for excellent technical assistance. This study was supported by the Deutsche Forschungsgemeinschaft (SFB 311), the Verband der Chemischen Industrie, and by a grant from the U.S. Department of Energy (to H.B.). Part of this work is contained in the M.D. thesis of M. Pinkernell.
References
- 1.Lakey J H, van der Goot F G, Pattus F. Toxicology. 1994;87:85–108. doi: 10.1016/0300-483x(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 2.Li J D, Carroll J, Ellar D J. Nature (London) 1991;343:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 3.Bhakdi S, Bayley H, Valeva A, Walev I, Walker B, Kehoe M, Palmer M. Arch Microbiol. 1996;165:73–79. doi: 10.1007/s002030050300. [DOI] [PubMed] [Google Scholar]
- 4.Liu C C, Persechini P M, Young J D. Immunol Rev. 1995;146:145–175. doi: 10.1111/j.1600-065x.1995.tb00688.x. [DOI] [PubMed] [Google Scholar]
- 5.Papini E, Sandona D, Rappuoli R, Montecucco C. EMBO J. 1988;7:3353–3359. doi: 10.1002/j.1460-2075.1988.tb03207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhakdi S, Tranum-Jensen J. Microbiol Rev. 1991;55:733–751. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker B, Braha O, Cheley S, Bayley H. Chem Biol. 1995;2:99–105. doi: 10.1016/1074-5521(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 8.Valeva A, Weisser A, Walker B, Kehoe M, Bayley H, Bhakdi S, Palmer M. EMBO J. 1996;15:1857–1864. [PMC free article] [PubMed] [Google Scholar]
- 9.Prendergast F G, Lu J, Callahan P J. J Biol Chem. 1983;258:7541–4078. [PubMed] [Google Scholar]
- 10.Valeva A, Palmer M, Hilgert K, Kehoe M, Bhakdi S. Biochim Biophys Acta. 1995;1236:213–218. doi: 10.1016/0005-2736(95)00060-g. [DOI] [PubMed] [Google Scholar]
- 11.Ward R J, Palmer M, Leonard K, Bhakdi S. Biochemistry. 1994;33:7477–7484. doi: 10.1021/bi00189a056. [DOI] [PubMed] [Google Scholar]
- 12.Walker B, Kasianowicz J, Krishnasastry M, Bayley H. Protein Eng. 1994;7:655–662. doi: 10.1093/protein/7.5.655. [DOI] [PubMed] [Google Scholar]
- 13.Walker B, Krishnasastry M, Bayley H. J Biol Chem. 1993;268:5285–5292. [PubMed] [Google Scholar]
- 14.Walker B, Krishnasastry M, Zorn L, Bayley H. J Biol Chem. 1992;267:21782–21786. [PubMed] [Google Scholar]
- 15.Palmer M, Weller U, Messner M, Bhakdi S. J Biol Chem. 1993;268:11963–11967. [PubMed] [Google Scholar]
- 16.Bhakdi S, Füssle R, Tranum-Jensen J. Proc Natl Acad Sci USA. 1981;78:5475–5479. doi: 10.1073/pnas.78.9.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouaux J E, Braha O, Hobaugh M R, Song L, Cheley S, Shustak C, Bayley H. Proc Natl Acad Sci USA. 1994;91:12828–12831. doi: 10.1073/pnas.91.26.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L, Hobaugh M R, Shustak C, Cheley S, Bayley H, Gouaux J E. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 19.Valeva A, Pongs J, Bhakdi S, Palmer M. Biochim Biophys Acta. 1997;1325:281–286. doi: 10.1016/s0005-2736(96)00266-0. [DOI] [PubMed] [Google Scholar]
- 20.Cormack B. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D M, Seidman J G, Smith J A, Struhl K, editors; Ausubel F M, Brent R, Kingston R E, Moore D M, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1987. , Chapter 8.5. [Google Scholar]
- 21.Jonas D, Schultheis B, Klas C, Krammer P H, Bhakdi S. Infect Immun. 1993;61:1715–1721. doi: 10.1128/iai.61.5.1715-1721.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhakdi S, Greulich S, Muhly M, Eberspacher B, Becker H, Thiele A, Hugo F. J Exp Med. 1989;169:737–754. doi: 10.1084/jem.169.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walev I, Martin E, Jonas D, Mohamadzadeh M, Müller-Klieser W, Kunz L, Bhakdi S. Infect Immun. 1993;61:4972–4979. doi: 10.1128/iai.61.12.4972-4979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer M, Jursch R, Weller U, Valeva A, Hilgert K, Kehoe M, Bhakdi S. J Biol Chem. 1993;268:11959–11962. [PubMed] [Google Scholar]
- 25.Walev I, Palmer M, Martin E, Jonas D, Weller U, Höhn-Bentz H, Husmann M, Bhakdi S. Microb Pathog. 1994;17:187–201. doi: 10.1006/mpat.1994.1065. [DOI] [PubMed] [Google Scholar]
- 26.Huang C, Mason J T. Proc Natl Acad Sci USA. 1977;75:308–310. doi: 10.1073/pnas.75.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodle M, Papahadjopoulos D. Methods Enzymol. 1989;171:193–217. doi: 10.1016/s0076-6879(89)71012-0. [DOI] [PubMed] [Google Scholar]
- 28.Olson F, Hunt C A, Szoka F C, Vail W J, Papahadjopoulos D. Biochim Biophys Acta. 1979;557:9–23. doi: 10.1016/0005-2736(79)90085-3. [DOI] [PubMed] [Google Scholar]
- 29.Oh K J, Zhan H, Cui C, Hideg K, Collier R J, Hubbell W L. Science. 1996;273:810–812. doi: 10.1126/science.273.5276.810. [DOI] [PubMed] [Google Scholar]
- 30.Walev I, Palmer M, Valeva A, Weller U, Bhakdi S. Infect Immun. 1995;63:1188–1194. doi: 10.1128/iai.63.4.1188-1194.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan B P. Curr Top Microbiol Immunol. 1992;178:115–140. doi: 10.1007/978-3-642-77014-2_8. [DOI] [PubMed] [Google Scholar]
- 32.Houle J J, Hoffmann E M, Esser A F. Blood. 1988;71:280–286. [PubMed] [Google Scholar]