Abstract
In the majority of cervical cancers, DNAs of high-risk mucosotpropic human papillomaviruses (HPVs), such as type 16, are maintained so as to express two viral proteins, E6 and E7, suggesting an essential importance to carcinogenesis. The high-risk HPV E6 proteins are known to inactivate p53 tumor suppressor protein but appear to have an additional, molecularly unknown function(s). In this study, we demonstrate that these E6 proteins can bind to the second PDZ domain of the human homologue of the Drosophila discs large tumor suppressor protein (hDLG) through their C-terminal XS/TXV/L (where X represents any amino acid, S/T serine or threonine, and V/L valine or leucine) motif. This finding is similar to the interaction between the adenomatous polyposis coli gene product and hDLG. E6 mutants losing the ability to bind to hDLG are no longer able to induce E6-dependent transformation of rodent cells. These results suggest an intriguing possibility that interaction between the E6 protein and hDLG or other PDZ domain-containing proteins could be an underlying mechanism in the development of HPV-associated cancers.
E6 and E7 genes of “high-risk” human papillomaviruses (HPVs), such as HPV16 and -18, have been called oncogenes (and their products, oncoproteins) for several reasons: (i) they are selectively retained and expressed in cervical cancer cells (1–4); (ii) the encoded proteins can interact with the so-called tumor suppressors, p53 and Rb family proteins, respectively, so as to abolish their function (5–7); and (iii) they can transform rodent cells (8, 9) and immortalize primary human epithelial cells (10–14). High risk HPV E6 proteins promote the ubiquitin-dependent proteolysis of p53 in cooperation with the cellular factor, E6AP (6, 7, 15–19). We and others have accumulated evidence indicating that they have p53-independent functions. HPV16 E6 transforms NIH 3T3 and 3Y1 cells but trans-dominant p53 mutants do not (8, 9). We have found that it can transform p53-deficient mouse fibroblasts (T.K. and M.I., unpublished data). In p53-deficient mice, HPV16 E6 inhibits apoptosis during lens development (20). HPV E6 proteins can bind a number of cellular proteins other than p53 in vitro (21). Indeed, a cDNA for a truncated form of a calcium-binding protein, ERC-55, named E6BP, was isolated with the yeast two-hybrid method using HPV16 E6 as “bait” and HeLa cells as the cDNA library source (22).
With essentially the same approach, a cDNA encoding a novel protein containing a PDZ domain has been isolated, and the domain has been found to be indispensable for the interaction (T.K. and M.I., unpublished data). PDZ domains, also known as DHR or GLGF motifs, were first noted as repeat sequences in the Drosophila septate junction protein DLG (the Drosophila discs large protein), the postsynaptic density protein PSD-95/SAP90, and the epithelial tight junction protein ZO-1 (23, 24), and have subsequently been identified in a diverse set of proteins that are typically associated with cell junctions. In Drosophila, mutation of the gene for DLG results in loosening of cell–cell contact and neoplastic proliferation of imaginal disc epithelial cells (25, 26). The presence of a PDZ domain in the novel E6-binding protein and the recently described PDZ-domain binding motif led us to note that the E6 proteins of high-risk mucosotropic HPVs, but not of other groups of HPVs, contain a conserved motif, XS/TXV/L (in single letter amino acid code, where X is any residue), at their C termini (Fig. 1). As the motif is the same or very similar to the C-terminal XS/TXV motif that has been found in a large number of ion channel and receptor molecules (27) and demonstrated by a crystallographic study to interact with the hydrophobic groove made by a PDZ domain (28), we hypothesized that high-risk HPV E6 proteins can bind PDZ domain-bearing proteins through their C-terminal motif. Because hDLG (the human homologue of the Drosophila discs large protein) is a tumor-suppressor protein in Drosophila, and hDLG is reported to bind to another tumor suppressor protein APC (the adenomatous polyposis coli gene product), we focused attention on hDLG as a putative E6 target.**
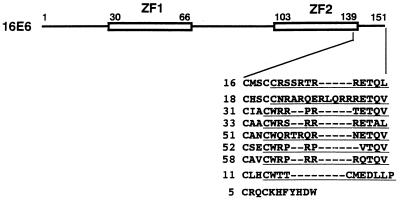
Figure 1.
An XS/TXV/L motif conserved specifically among E6 proteins of high-risk mucosotropic HPVs. HPV16 E6 polypeptides and the C-terminal region sequence alignment of several HPV E6 proteins are represented schematically. The two zinc-finger domains (ZF1 and ZF2) are indicated with the position numbers of the amino acid residues. The whole sequences of HPV E6 proteins were aligned by clustalw (version 1.6; Human Genome Center, Baylor College of Medicine), but only the results for the C-terminal region are presented. The sequences used as the C-terminal oligopeptides (CTs) are underlined. Arabic numbers prior to the amino acid sequences shown as single letter abbreviations indicate the HPV types.
MATERIALS AND METHODS
Plasmids.
Segments of HPV16 E6 gene (16E6) that have a point mutation at the splicing donor site (16E6SD) were described (9). HPV16 E6 genes for the C-terminal mutants were generated by replacing a PvuII–BamHI short segment containing the C-terminal (151st) and the termination codons of 16E6 or 16E6SD with appropriate double-stranded synthetic oligonucleotides. Other E6 gene segments were described (9). These E6 segments containing BamHI cohesive ends and a small leader sequence, ACC, for efficient translation (20) flanking the initiation codon, were inserted in pGEM (Promega) or pBluescript (Stratagene) for in vitro translation and in a retrovirus plasmid, pLRHL, constructed by replacing the neomycin resistant gene of pLRNL (29) with the hygromycin-B resistance gene for expression in rodent cells. Double-stranded synthetic oligonucleotides containing the coding sequence for the myc-epitope tag (30) followed by a BamHI recognition site were inserted into NotI–BstX1 sites of pEF-BOS (31), giving pEF-MYC. The same E6 gene fragments as above except for lacking the translation initiation codon were inserted into the BamHI site of pEF-MYC so that the amino-terminally myc-tagged E6 proteins could be expressed under the control of the human polypeptide chain elongation factor 1α promoter. The resultant plasmids were confirmed to have the correctly ligated segments by sequencing.
Transformation Assays.
Various E6 genes cloned in pLRHL were transiently transfected into PA317 cells by electroporation, and after 72 h conditioned medium was used for infection of 3Y1. Infection, drug selection, and culture conditions were as described (32, 33). Morphological transformation could be detected as early as 2 weeks after infection. Various E6 genes in pLRHL were transfected into ψ2 cells using calcium phosphate as described (32). About 2 weeks after selection with hygromycin-B (100 μg/ml), several hundred drug-resistant colonies were pooled and used for injection. Tumorigenicity was tested by injecting 2 × 105 cells subcutaneously into BALB/c, (nu/nu) mice at 4 weeks of age and measuring the size of the resulting tumors at least twice a week for 36 days. In each case pooled cells were injected at two sites per mouse.
Protein Interaction Assays.
Various [35S]cysteine-labeled E6 proteins as well as hDLG were synthesized using the in vitro transcription and translation-coupled reticulocyte lysate system (Promega). Plasmids for maltose-binding protein (MBP) fusion proteins were constructed by inserting various E6 segments into pMALcRI (New England Biolabs), and those for glutathione S-transferase (GST) fusion proteins were as described (33). MBP and GST fusion proteins were isolated as detailed earlier (34, 35). Those proteins immobilized to amylose- and glutathione-Sepharose, respectively, were preincubated in Nonidet P-40 lysis buffer [1% Nonidet P-40/150 mM NaCl/50 mM Tris⋅HCl, pH 7.5)] containing 2% BSA for at least 10 min, mixed with 4–20 μl of the programmed reticulocyte lysates containing in-vitro translated (IVT) products, and, in the case of competition assays, indicated amounts of oligopeptides corresponding to the C-terminal regions of HPV E6 proteins for 2 h at 4°C and then washed with 800 μl of Nonidet P-40 lysis buffer five times. Samples were resolved by SDS/PAGE. All the gels were first subjected to Coomassie staining so as to confirm the comparable amounts of MBP or GST fusion proteins were loaded on gels, and analyzed with a Bio-Imaging system BAS2000 (Fuji) for visualizing the captured proteins.
Immunoprecipitation and Immunoblotting.
pEF-MYC-based plasmid DNAs were introduced into monkey COS cells by electroporation. Forty-eight hours later the cells were lysed in Nonidet P-40 lysis buffer containing 1 mM phenylmethylsulfonyl fluoride, leupeptine (10 μg/ml), and aprotinin (10 μg/ml), and the lysates were incubated with anti-myc epitope antibody (9E10) for 1 h at 4°C. The immunocomplexes were adsorbed to protein G-Sepharose 4B (Pharmacia) and washed with Nonidet P-40 lysis buffer five times. Samples were resolved by SDS/PAGE. Proteins separated in an SDS polyacrylamide gel were blotted to a polyvinylidene difluoride membrane (Immobilon-P, Millipore). The membrane was cut into two pieces so that the upper should be blotted with hDLG and the lower with myc-tagged E6 proteins. The upper blot was incubated with rabbit anti-hDLG antiserum (33) at 1:500 dilution and the lower with purified rabbit anti-myc tag antibodies (Medical and Biological Laboratories, Nagoya, Japan) at 1 μg/ml for 1 h at room temperature. The blots were then probed with peroxidase-labeled goat anti-rabbit IgG (Zymed) and visualized with the chromogenic substrate system (Immunostain HRP kit, Konica, Tokyo).
Infection of Primary Human Mammary Epithelial Cells.
Plasmid DNA was transiently transfected into PE501 cells by electroporation, and after 72 h conditioned medium was used for infection of PG13 (36) followed by selection in hygromycin B (0.4 mg/ml) to establish clonal lines producing amphotropic recombinant retroviruses containing various E6 genes. Human mammary epithelial cells were infected with these recombinant retroviruses and selected in hygromycin-B (20 μg/ml) for 4 days. The resultant colonies were pooled and propagated for further analysis. The exponentially growing cultures of these pooled cells 13 days after infection were treated with or without 2.5 μM actinomycin D. After 24 h cellular proteins were extracted and subjected to immunoblotting for p53 and p21 as described (37).
RESULTS AND DISCUSSION
Interactions of E6 Proteins of High Risk Mucosotropic HPVs with hDLG.
First to examine whether various E6 proteins can bind to hDLG, various MBP E6 proteins immobilized on amylose Sepharose beads were mixed with IVT-hDLG and sedimented. IVT-hDLG could efficiently bind to E6 proteins of HPV16 and -18 but not of HPV11 and -5 (Fig. 2A). In a reciprocal experiment using various IVT-E6 proteins and a GST-hDLG fusion protein immobilized on glutathione Sepharose beads supported the above results (Fig. 2B). Thus E6 proteins of high-risk mucosotropic HPVs specifically bind to hDLG. The binding of hDLG to MBP-16E6 and -18E6 was inhibited by coincubation with a series of oligopeptides corresponding to the C-terminal amino acid residues of high-risk HPV E6 proteins but not with that of 16E6 lacking the terminal L or 11E6 (Figs. 1 and 2C; data not shown for MBP-18E6). These results not only indicate the importance of the C-terminal region in determining the binding specificity, but also imply that the E6 proteins of high-risk HPVs other than type 16 and 18 can bind hDLG through the common C-terminal motif.
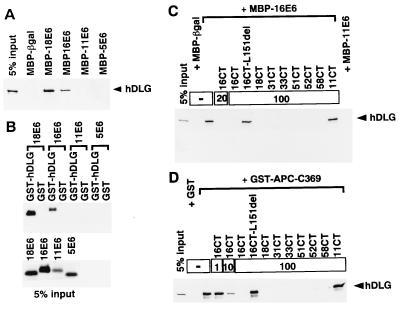
Figure 2.
Interactions of E6 proteins of high-risk mucosotropic HPVs as well as APC with hDLG, and their inhibition by C-terminal oligopeptides (CTs) corresponding to high-risk mucosotropic HPV E6 proteins (see Fig. 1). (A) Radiolabeled IVT-hDLG was incubated with various MBP-E6 proteins or with MBP-βgal (β-galatosidase of Escherichia coli), and the captured proteins together with 5% equivalents of the input IVT-hDLG were resolved by SDS/PAGE. Autoradiographic electropherograms of the areas where IVT-hDLG proteins are expected to migrate are shown. (B) IVT-E6 proteins of various HPVs were mixed with GST or GST-hDLG protein-Sepharose (Upper), and the captured proteins were treated as for A. (Lower) Autoradiogram of 5% equivalents of the input IVT-E6s. (C) Mixtures of IVT-hDLG and MBP-16E6-Sepharose were coincubated with the indicated amounts (μg) of various CTs, and the captured proteins were treated as in A. (D) Same as for C except that mixtures of GST-APC-C369 protein-Sepharose and IVT-hDLG were used.
The Oligopeptides Corresponding to the C Termini of High-Risk HPV E6 Proteins Interfere with Binding Between APC and hDLG.
If the binding could occur in vivo, it is likely that such interaction would prevent DLGs from binding to cellular XS/TXV motif-bearing proteins such as APC. This notion is in line with the results of an in vitro experiment in which binding of hDLG to a GST-fused C-terminal domain of APC, GST-APC-C369 (33), was preferentially disturbed by coincubation with the above-described oligopeptides corresponding to the C termini of high-risk HPV E6 proteins (Figs. 1 and 2D). These results indicate that the common motif shared by APC and the E6s determines the binding specificity to hDLG.
The Second PDZ Domain of hDLG Is Necessary for Binding to HPV16 E6 Protein.
We next delimited the segment of hDLG necessary for binding to 16E6 (Fig. 3). GST fusion proteins containing only PDZ domains R1, -2, and -3 (R123) captured IVT-16E6 as efficiently as those containing the full-length hDLG. Proteins containing only the first and the second domains (R12) or only the second domain (R2) bound less efficiently, and those containing only the third domain (R3) showed very weak binding (Fig. 3 A and B). MBP-16E6-bound truncated hDLG containing only three PDZ domains or the full-length hDLG efficiently, whereas the hDLG segment ranging from the N terminus to the first PDZ domain was not bound (Fig. 3 A and C). MBP-16E6-bound hDLG segment containing only the first and the second domains (R12) but with reduced efficiency. These results indicate that the second PDZ domain is essential for efficient binding to 16E6, as in the case of APC binding to hDLG (33), and that the third domain might also contribute to the interaction to some extent.
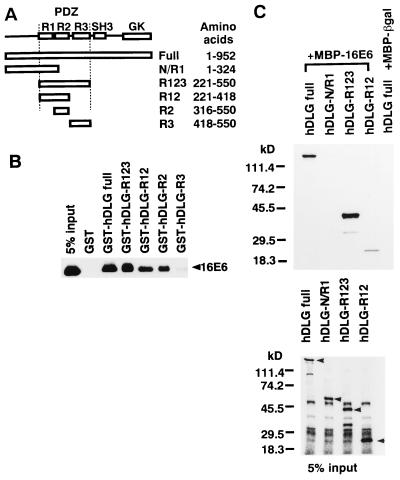
Figure 3.
Determination of the hDLG protein segment necessary for binding to HPV16 E6 protein. (A) Schematic representation of hDLG and its deletion mutants (33). The locations of the three repeats (R1, R2, and R3) of the PDZ domain (PDZ), a Src homology 3 (SH3) domain, and a putative guanylate kinase domain (GK) are indicated. (B) Various segments of hDLG shown in A were prepared as GST-fused forms immobilized to glutathione-Sepharose, and they or GST were mixed with IVT-16E6. The captured proteins were treated as in Fig. 2A. (C) Mixtures of MBP-16E6-Sepharose and various segments of hDLG (see A) as IVT-forms were treated as in Fig. 2A Upper. The lower panel shows 5% equivalents of IVT products similarly treated. Arrowheads indicate positions of the bands corresponding to the specific IVT products.
The C-Terminal Motif of HPV16 E6 Protein Is Critical for Interaction with hDLG.
To further examine the critical role of the C-terminal region of 16E6, a series of mutants in its XS/TXV/L motif, namely ETQL (Glu-Thr-Gln-Leu), were prepared and examined for their ability to bind hDLG in vitro (Fig. 4A, Table 1). The results demonstrated that the substitution of the last residue, L (Leu), with another hydrophobic residue, I (Ile), as well as V (Val), does not exert any significant effect, but that substitution with P (Pro), deletion of the terminal L, and addition of P after the L completely abolish the binding ability. Furthermore, addition of the most common last four amino acid sequence among high-risk HPVs, ETQV (Glu-Thr-Gln-Val) to the one amino acid-deleted 16E6 restored the binding ability. These results fit well with the hypothesis that the C-terminal four amino acid residues are important for the binding.
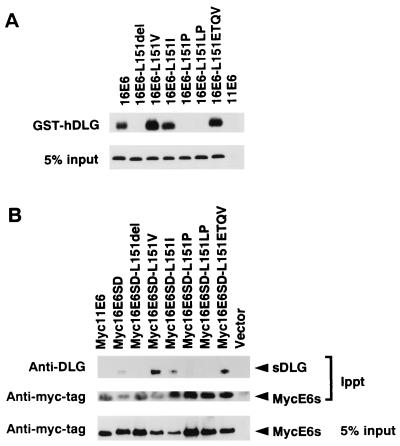
Figure 4.
Role of the C-terminal region of HPV16 E6 protein in its interaction with hDLG. (A) Mutant 16E6s whose 151st (C-end) amino acid residue was modified, and 11E6 as a control, were prepared as IVT forms mixed with GST-hDLG-Sepharose, and the captured proteins were treated as for Fig. 2A. The autoradiographic profiles of the areas where IVT-E6 mutant proteins were expected to migrate are shown. Five percent equivalents of the input IVT-E6s are also illustrated. (B) The mutant 16E6s examined in A were tagged with a myc-epitope and expressed in COS cells because high-avidity antibodies against 16E6 are not available. Cell lysates were then subjected to immunoprecipitation with anti-myc epitope antibody (9E10), and the precipitates were fractionated by SDS/PAGE and immunoblotted with anti-hDLG (33) and rabbit anti myc tag antibodies. Five percent equivalents of the lysates used for the immunoprecipitation were also similarly treated with the rabbit anti-myc tag antibodies for comparison.
Binding of HPV16 E6 with hDLG in COS Cell Lysate.
To test whether the same binding specificity as observed in vitro also takes place in vivo, mutant 16E6s with a myc epitope-tag at the N terminus and an additional point mutation at the splicing donor site to assure higher expression of the full-length E6 (9) were expressed in COS cells, and the cell lysates were exposed to anti-myc tag antibodies. Significant amounts of simian DLG were coprecipitated with 16E6 and the mutant 16E6s shown to bind to hDLG in vitro but not with the other mutant 16E6s or 11E6 (Fig. 4B, Table 1). It is worthy to note that the two mutants ending with Val, L151V and L151ETQV, which conform more common C-terminal motif among high-risk HPV E6 proteins, interacted significantly better with human or simian DLG than wild-type 16E6 (Fig. 4 A and B). Taking account of the fact that 16E6 has not been identified in complexes with any of the previously reported E6-binding proteins, p53, E6AP, and E6BP, in vivo, these results are particularly worthy of note, providing more direct support for the proposal that E6 proteins of high-risk mucosotropic HPVs bind to mammalian DLG in vivo through the C-terminal motif.
The C-Terminal Motif of HPV16 E6 Protein Is Necessary for Morphological Transformation of a Rat Fibroblastic Cell Line.
If the high-risk HPV E6 proteins target hDLG and/or some other PDZ domain-bearing proteins, which are typically localized at cell junctions, it seems likely that they would impair the growth arrest induced by cell–cell contact in vivo, as assumed to be the case of Drosophila with a mutant DLG (25, 26). For this reason, and because 16E6 can transform 3Y1 cells despite its failure to inactivate their p53 activity (9), we tested whether these mutant 16E6s could transform these cells, which show strong growth arrest when they reach confluence and form monolayers with a cobble-stone appearance (38). 16E6 and the mutant 16E6s shown to bind to hDLG, but not the other mutant 16E6s or 11E6, transformed 3Y1 cells so that the confluent cell sheets showed multilayered swirl forming arrays of spindle-like cells rather than the monolayers (39) (Table 1, photographs not shown). The results indicate that interaction of 16E6 through its C-terminal motif with PDZ domain-bearing proteins like hDLG is necessary for transformation.
The C-Terminal Motif of HPV16 E6 Protein Is Necessary for Tumorigenic Transformation of a Mouse Fibroblastic Cell Line.
Interestingly, HPV16 E6, but not E7, can endow tumorigenicity on ψ2 cells, which correlates with the expression level of the full-length E6 (32). We therefore tested representative mutant 16E6s for this ability (Table 1). ψ2 cells introduced with the mutant 16E6 shown to bind to hDLG and transform 3Y1 cells, as well as 16E6 itself, gave rise to tumors in nude mice as early as 16 days after injection, whereas no lesions were observed in those introduced with the other mutants or 11E6 and the vector alone, after even 36 days injection. These mutants retained the ability to target p53 for degradation in human mammary epithelial cells (data not shown), consistent with another report indicating that the C-terminal 12 amino acids of 16E6 are not required for p53 degradation or abrogation of actinomycin D-induced growth arrest (40). Taking into account these facts together with the inability of 16E6 to inhibit the p53 function in 3Y1 cells (9), the present results can be interpreted as indicating that interaction between E6 and PDZ domain-bearing protein(s) is crucial for E6 transformation independent of effects on p53. Because the mammalian homologue of DLG is ubiquitously expressed in many organs in vivo and in cell lines including HeLa and NIH 3T3 (21) from which ψ2 is derived, and we have detected the rat homologue of DLG in 3Y1 cells by Western blot analysis (data not shown), the DLG itself could be the target of the E6 protein for transformation and oncogenesis.
Mutation of APC is thought to be an early event in tumorigenesis of colorectal tumors (41), and APC can bind to hDLG (33) so as to lead cell cycle arrest (T.A., unpublished data). Our finding of an interaction between the E6 proteins and hDLG not only suggests the possibility that E6 proteins might disturb and/or modify such signal transduction pathways depending on the motif domain interaction, but also implies the intriguing possibility that there might be a common or similar mechanism underlying the development of colorectal and HPV-associated cancers. In addition to the interaction with the XS/TXV motif, PDZ domains also form apparently specific homophilic associations with other PDZ domains (38), thereby apparently contributing to many signal transduction pathways, including that leading to apoptosis (42). A crystallographic study has suggested that each PDZ domain recognizes a specific XS/TXV motif, and that the specificity is, at least in part, determined by the first X residue in the motif (28). More recently, the optimal sequences recognized by distinct PDZ domains were reported by using oligopeptide library, and the C-terminal sequences of high-risk HPV E6 proteins match well to those reported to be recognized by the mouse homologue of DLG (43). These studies further support our results and suggest it likely that the E6 proteins can bind to only a limited set of PDZ domain-bearing proteins including hDLG, though one cannot rule out the possibility that there might be a PDZ domain-bearing protein(s) more important than hDLG as the E6 target(s). PTPbas/FAP-1 known to bind Fas antigen and modulate apoptosis (42) might be a such candidate because the E6 proteins can inhibit apoptosis in not only p53-dependent but also p53-independent manner (44). Recently a paper describing in vitro interaction of hDLG with human T cell leukemia virus type 1 Tax and HPV18 E6 proteins as well as in vivo interaction between hDLG and adenovirus type 9 E4 ORF1 protein was published (45). Our results with HPV E6 proteins are consistent with this study. Clearly, the potential significance of interactions between these viral oncoproteins and the PDZ domain-bearing proteins in transformation in vitro and carcinogenesis in vivo, as well as virus production, warrants further attention.
Table 1.
Transforming activity of HPV16 E6 mutants
| Designation | C end | Binding to DLG | Transformation of 3Y1 cells | Tumorigenicity of ψ2 cells† |
|---|---|---|---|---|
| 16E6 | ETQL | + | + | 9/10† (16-36‡) |
| 16E6L151del | ETQ | − | − | 0/10§ |
| 16E6L151V | ETQV | + | + | 7/10 (16-36) |
| 16E6L151I | ETQI | + | + | NT |
| 16E6L151P | ETQP | − | − | 0/10§ |
| 16E6L151LP | ETQLP | − | − | NT |
| 16E6L151ETQV | ETQETQV | + | + | NT |
| 18E6 | ETQV | + | + | NT |
| 11E6 | DLLP | − | − | 0/10‡ |
NT, not tested.
Summarized data from Fig. 4.
Shown are the number of tumors (larger than 5 mm in diameter) per number of injected sites. Each mouse was injected at two sites. The minimal time (days, in parentheses) to produce apparent tumors of at least 5 mm in diameter.
No tumors were observed 36 days after injection.
Acknowledgments
We thank M. Noda, R. Haruta, K Irie, T. Yoshida, and C. Yamada for technical assistance, and Denise A. Galloway, Scott A. Foster, and Harald zur Hausen for reading the manuscript. We also thank Denise A. Galloway for letting us conduct a part of this study in her laboratory. This work is supported in part by grants-in-aid for Cancer Research from the Ministry of Education, Science and Culture, Japan.
ABBREVIATIONS
- HPV
human papillomavirus
- hDLG
the human homologue of the Drosophila discs large protein
- DLG
the Drosophila discs large protein
- APC
the adenomatous polyposis coli gene product
- GST
glutathione S-transferase
- MBP
maltose binding protein
- IVT
in vitro translated
Footnotes
The major part of the present study was presented orally at the 15th International Papillomavirus Conference in Brisbane, Australia 1996.
References
- 1.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smotkin D, Wettstein F O. Proc Natl Acad Sci USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Nature (London) 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 4.Lehn H, Krieg P, Sauer G. Proc Natl Acad Sci USA. 1985;82:5540–5544. doi: 10.1073/pnas.82.16.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyson N, Howley P M, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 6.Werness B A, Levine A J, Howley P M. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 8.Sedman S A, Hubbert N L, Vass W C, Lowy D R, Schiller J T. J Virol. 1992;66:4201–4208. doi: 10.1128/jvi.66.7.4201-4208.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiyono T, Hiraiwa A, Ishii S, Takahashi T, Ishibashi M. J Virol. 1994;68:4656–4661. doi: 10.1128/jvi.68.7.4656-4661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halbert C L, Demers G W, Galloway D A. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Band V, De Caprio J A, Delmolino L, Kulesa V, Sager R. J Virol. 1991;65:6671–6676. doi: 10.1128/jvi.65.12.6671-6676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shay J W, Wright W E, Brasiskyte D, Van der Haegen B A. Oncogene. 1993;8:1407–1413. [PubMed] [Google Scholar]
- 15.Huibregtse J M, Scheffner M, Howley P M. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 18.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crook T, Tidy J A, Vousden K H. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 21.Keen N, Elston R, Crawford L. Oncogene. 1994;9:1493–1499. [PubMed] [Google Scholar]
- 22.Chen J J, Reid C E, Band V, Androphy E J. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 23.Cho K O, Hunt C A, Kennedy M B. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 24.Gomperts S N. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 25.Woods D F, Bryant P J. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 26.Lue R A, Marfatia S M, Branton D, Chishti A H. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 28.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 29.Emi N, Friedmann T, Yee J K. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cravchik A, Matus A. Gene. 1993;137:139–143. doi: 10.1016/0378-1119(93)90262-2. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue T, Kyo S, Kiyono T, Ishibashi M, Ishiwatari H, Hwang Y I, Yutsudo M, Hakura A. Jpn J Cancer Res. 1994;85:357–363. doi: 10.1111/j.1349-7006.1994.tb02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg G H, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 34.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 35.Blondel A, Bedouelle H. Eur J Biochem. 1990;193:325–330. doi: 10.1111/j.1432-1033.1990.tb19341.x. [DOI] [PubMed] [Google Scholar]
- 36.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster S A, Galloway D A. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- 38.Brenman J E, Chao D S, Gee S H, McGee A W, Craven S E, Santillano D R, Wu Z, Huang F, Xia H, Peters M F, Froehner S C, Bredt D S. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 39.Hiraiwa A, Kiyono T, Segawa K, Utsumi K R, Ohashi M, Ishibashi M. Virology. 1993;192:102–111. doi: 10.1006/viro.1993.1012. [DOI] [PubMed] [Google Scholar]
- 40.Foster S A, Demers G W, Etscheid B G, Galloway D A. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogelstein B, Kinzler K W. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 42.Sato T, Irie S, Kitada S, Reed J C. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 43.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 44.Pan H, Griep A E. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 45.Lee S S, Weiss R S, Javier R T. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]