Abstract
Four tra delta FargG+ plasmids, derived from matings between Hfr AB312 and a recA recipient, have been shown to have deletions of at least 50% of the F genome, including the region in which the tra genes map. The mutant plasmids do contain the F genes required for plasmid maintenance. Correlations can be made between, on the one hand, the F genes present on the tradelta F' plasmids and the F genes transferred early by an Hfr donor, and, on the other hand, the F genes deleted from the tradelta F' plasmids and the F genes transferred late by an Hfr donor. A biased representation of proximally and distally transferred chromosomal markers among the tradelta F' elements was also demonstrated. Taken Taken together, the asymmetrical representation of Hfr genes and the cis dominance of the Tra phenotype of these mutants can best be explained by the hypothesis that the tradelta F' plasmids are formed by repliconation of the transferred exogenote in a recA recipient.
Full text
PDF

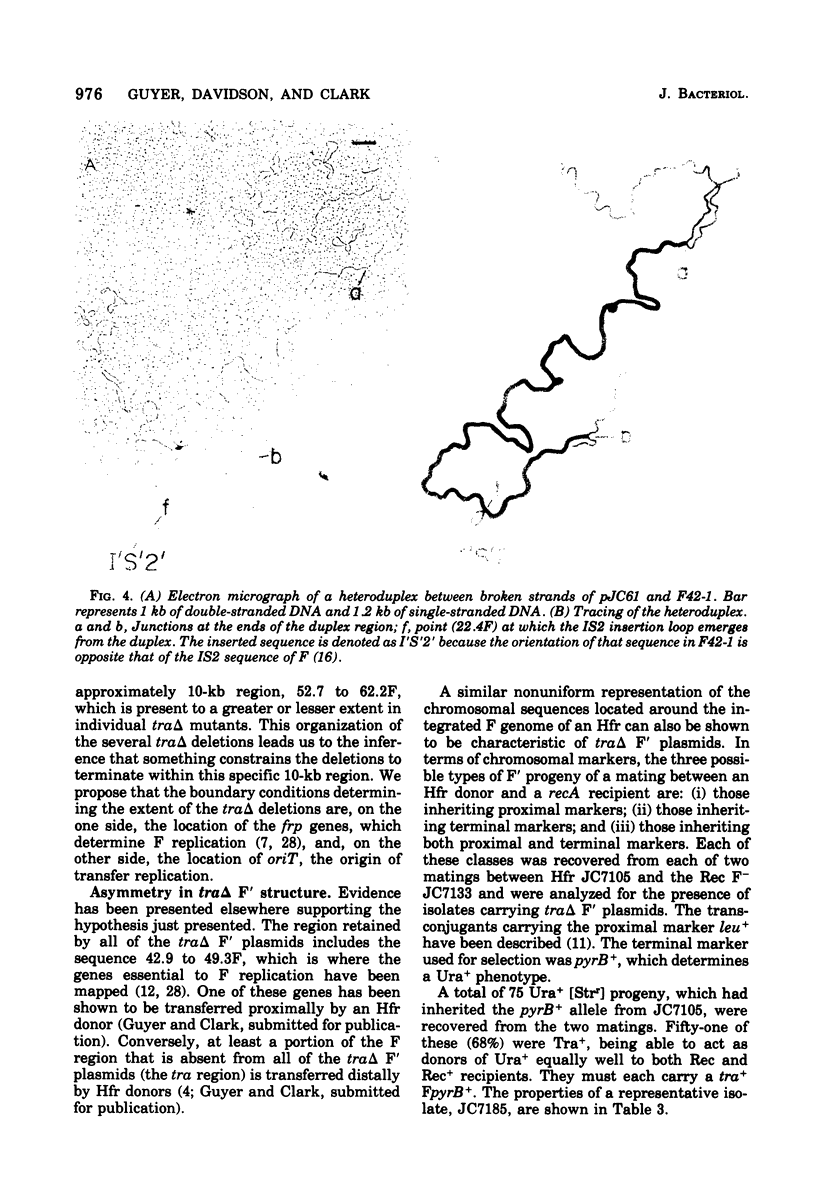
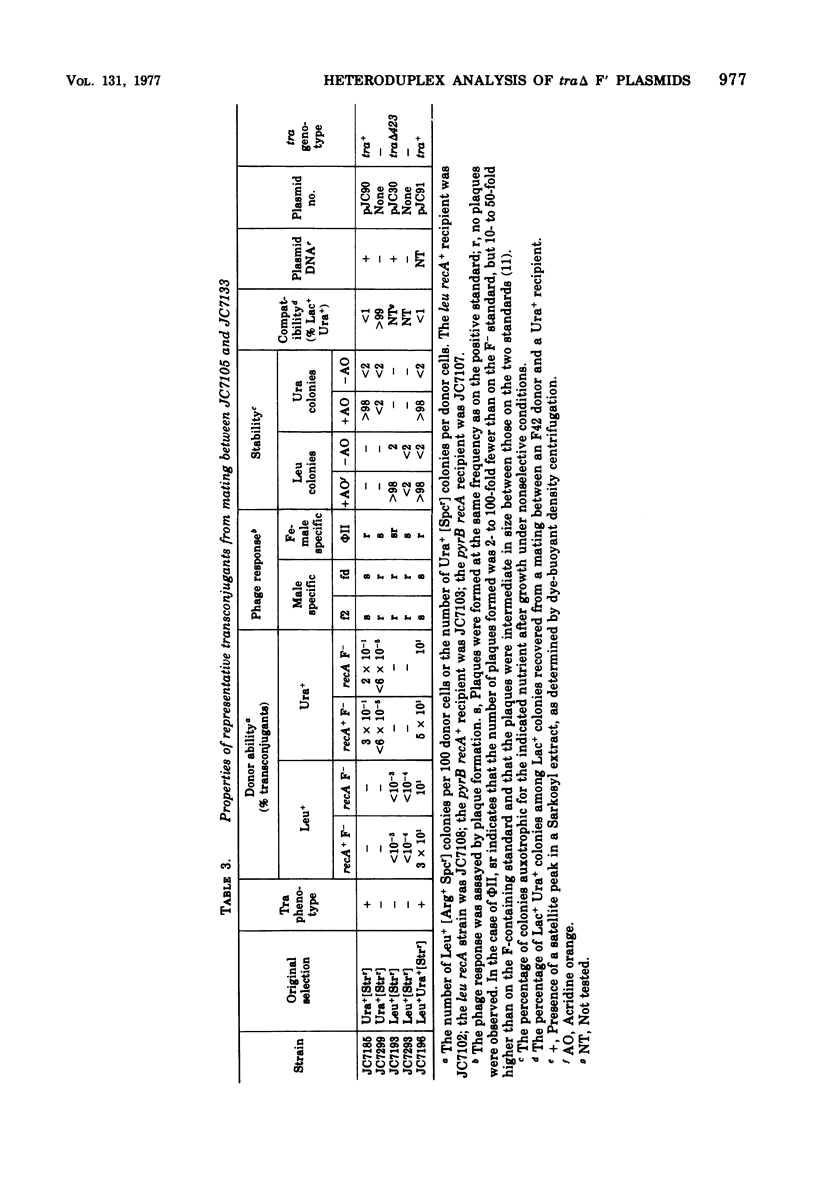
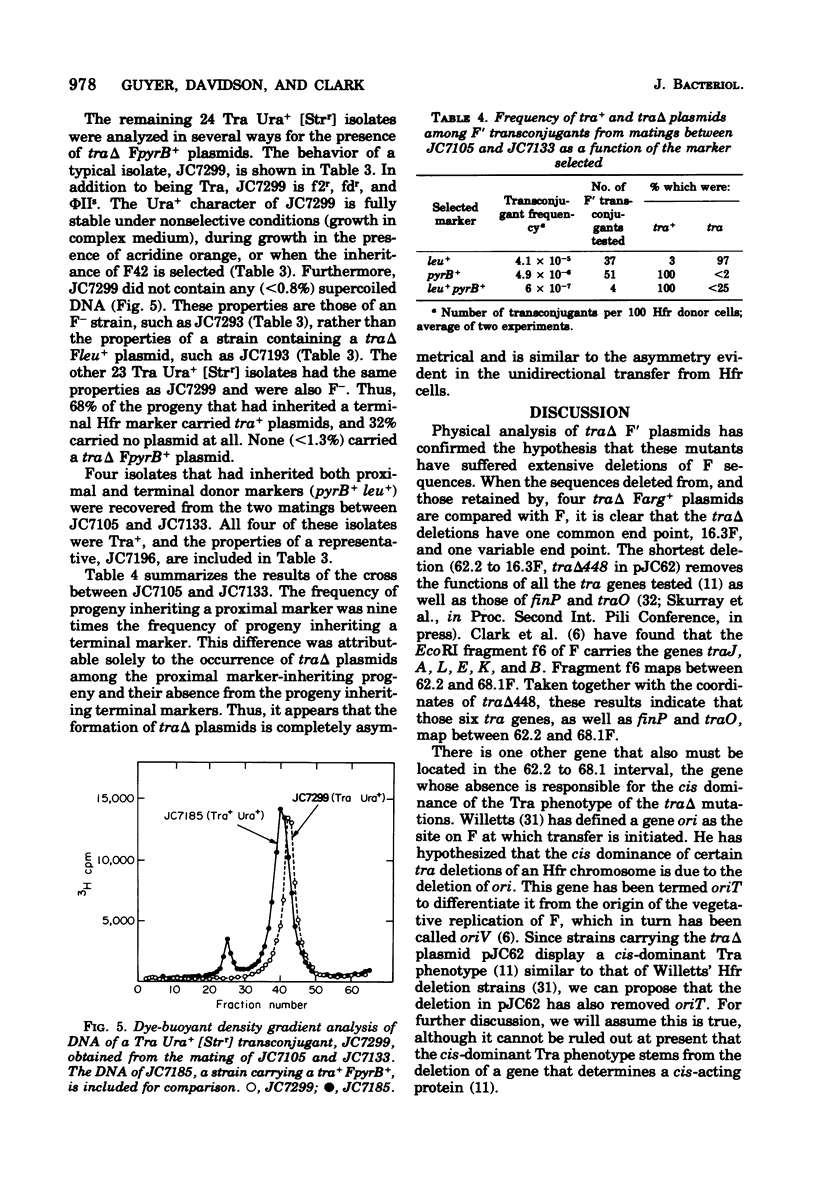


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M. Genetics of the F sex factor in enterobacteriaceae. Curr Top Microbiol Immunol. 1973;60:79–123. doi: 10.1007/978-3-642-65502-9_3. [DOI] [PubMed] [Google Scholar]
- Anthony W. M., Deonier R. C., Lee H. J., Hu S., Otsubo E., Davidson N., Broda P. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. IX. Note on the deletion mutant of F, F delta(33-43). J Mol Biol. 1974 Nov 15;89(4):647–650. doi: 10.1016/0022-2836(74)90041-2. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub R., Figurski D., Helinski D. R. Bidirection replication from a unique origin in a mini-F plasmid. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1138–1141. doi: 10.1073/pnas.74.3.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiandt M., Szybalski W., Malamy M. H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119(3):223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- Guyer M. S., Clark A. J. cis-Dominant, transfer-deficient mutants of the Escherichia coli K-12 F sex factor. J Bacteriol. 1976 Jan;125(1):233–247. doi: 10.1128/jb.125.1.233-247.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Figurski D., Davidson N. Electron microscope study of a plasmid chimera containing the replication region of the Escherichia coli F plasmid. J Bacteriol. 1976 Aug;127(2):988–997. doi: 10.1128/jb.127.2.988-997.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmuth R., Achtman M. Operon structure of DNA transfer cistrons on the F sex factor. Nature. 1975 Oct 23;257(5528):652–656. doi: 10.1038/257652a0. [DOI] [PubMed] [Google Scholar]
- Hirsch H. J., Starlinger P., Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119(3):191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- Hu S., Ohtsubo E., Davidson N. Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. J Bacteriol. 1975 May;122(2):749–763. doi: 10.1128/jb.122.2.749-763.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOB F., ADELBERG E. A. Transfert de caractéres génétiques par incorporation au facteur sexuel d'Escherichia coli. C R Hebd Seances Acad Sci. 1959 Jul 6;249(1):189–191. [PubMed] [Google Scholar]
- Johnson E. M., Craig W. G., Jr, Wohlhieter J. A., Lazere J. R., Synenki R. M., Baron L. S. Conservation of Salmonella typhimurium deoxyribonucleic acid in partially diploid hybrids of Escherichia coli. J Bacteriol. 1973 Aug;115(2):629–634. doi: 10.1128/jb.115.2.629-634.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters M. Strains of Escherichia coli diploid for the chromosomal origin of DNA replication. Mol Gen Genet. 1975 Dec 30;143(1):105–111. doi: 10.1007/BF00269427. [DOI] [PubMed] [Google Scholar]
- Nathans D., Smith H. O. Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. Extrachromosomal inheritance in bacteria. Bacteriol Rev. 1969 Jun;33(2):210–263. doi: 10.1128/br.33.2.210-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo E., Deonier R. C., Lee H. J., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. IV. The F sequences in F14. J Mol Biol. 1974 Nov 15;89(4):565–584. doi: 10.1016/0022-2836(74)90036-9. [DOI] [PubMed] [Google Scholar]
- Santos D. S., Palchaudhuri S., Maas W. K. Genetic and physical characteristics of an enterotoxin plasmid. J Bacteriol. 1975 Dec;124(3):1240–1247. doi: 10.1128/jb.124.3.1240-1247.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaife J. Episomes. Annu Rev Microbiol. 1967;21:601–638. doi: 10.1146/annurev.mi.21.100167.003125. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Skurray R. A., Guyer M. S., Timmis K., Cabello F., Cohen S. N., Davidson N., Clark A. J. Replication region fragments cloned from Flac+ are identical to EcoRI fragment f5 of F. J Bacteriol. 1976 Sep;127(3):1571–1575. doi: 10.1128/jb.127.3.1571-1575.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E. M., Pittard J. Conjugation in Escherichia coli: failure to confirm the transfer of part of sex factor at the leading end of the donor chromosome. J Bacteriol. 1972 May;110(2):516–522. doi: 10.1128/jb.110.2.516-522.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S. Location of the origin of transfer of the sex factor F. J Bacteriol. 1972 Nov;112(2):773–778. doi: 10.1128/jb.112.2.773-778.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Maule J., McIntire S. The genetic locations of traO, finP and tra-4 on the E. coli K12 sex factor F. Genet Res. 1975 Dec;26(3):255–263. doi: 10.1017/s0016672300016050. [DOI] [PubMed] [Google Scholar]
- Willetts N. The genetics of transmissible plasmids. Annu Rev Genet. 1972;6:257–268. doi: 10.1146/annurev.ge.06.120172.001353. [DOI] [PubMed] [Google Scholar]






