Abstract
The panneural protein Prospero is required for proper differentiation of neuronal lineages and proper expression of several genes in the nervous system of Drosophila. Prospero is an evolutionarily conserved, homeodomain-related protein with dual subcellular localization. Here we show that Prospero is a sequence-specific DNA-binding protein with novel sequence preferences that can act as a transcription factor. In this role, Prospero can interact with homeodomain proteins to differentially modulate their DNA-binding properties. The relevance of functional interactions between Prospero and homeodomain proteins is supported by the observation that Prospero, together with the homeodomain protein Deformed, is required for proper regulation of a Deformed-dependent neural-specific transcriptional enhancer. We have localized the DNA-binding and homeodomain protein-interacting activities of Prospero to its highly conserved C-terminal region, and we have shown that the two regulatory capacities are independent.
The neuronal precursor gene prospero (pros) encodes the neuronal specific protein Prospero (Pros) expressed in most or all neuronal lineages in Drosophila embryos (1–3). In the central nervous system, Pros is initially expressed in neuroblasts, where it is localized to the basal pole of the cell membrane (4–6). After the division of neuroblasts, Pros becomes localized to the nuclei of ganglion mother cells (GMC) (1–3), which subsequently divide to give rise to two terminally differentiated cells, and Pros remains nuclear in all cells expressing it. In the peripheral nervous system Pros shows a similar pattern of localization. It is initially associated with the cell membrane of sensory organ precursors (5), but then becomes localized to the nuclei of all subsequent cells in which it is expressed (2, 5).
Pros defines an evolutionarily conserved class of atypical homeodomain proteins with homologues in Caenorhabditis elegans, chicken, mouse, and human (7–10). Atypical homeodomain proteins are structurally homologous to typical Antennapedia class homeodomain proteins. However, their homeodomains display significant variation at the primary sequence level among themselves as well as in comparison with typical homeodomain proteins (11). In addition, atypical homeodomain proteins have extra amino acids inserted between helices 1 and 2 and/or helices 2 and 3. The amino acid sequence homology between Pros and typical homeodomain proteins is highest in the third helix of the predicted Pros homeodomain, where all consensus amino acids are conserved (1–3). In contrast, some highly conserved residues of the first and second helices are altered in Pros (1, 3, 11). Within the Pros family of proteins, there is extensive homology between Pros and its C. elegans, chicken, mouse, and human homologues extending over the 160 C-terminal amino acids of the four proteins (7–10). In this region, which includes the putative homeodomains of the Drosophila, C. elegans, and chicken proteins, the amino acid identity is between 58% (Drosophila and chicken; Drosophila and mouse) and 69% (Drosophila and C. elegans). A conserved region located C-terminal of the homeodomain has been called the Pros domain (8).
pros loss of function mutations result in the transcriptional misregulation of several genes, including the panneural neuronal precursor genes deadpan (dpn) and asense (ase) (2) and the neuronal subset-specific genes even-skipped (eve), fushi tarazu (ftz), and engrailed (en) (12). This regulatory defect is associated with severe defects in axonal outgrowth and glial differentiation (2, 12–14).
The nuclear localization of Pros, combined with the conserved homeodomain and the requirement of Pros function for the proper expression of several genes, indicates that Pros may function as a nervous system-specific DNA-binding transcription factor (1–3). Pros differentially regulates multiple genes during neuronal lineage development, raising the possibility that it cooperates with other transcription factors. Here we show that Pros is a sequence-specific DNA-binding protein that can act as a transcription factor. The C-terminal 236 amino acids of the protein, including the highly conserved Pros domain, are sufficient to mediate both DNA binding and some transcriptional activities. We also show that Pros differentially modulates the activities of several homeodomain proteins. This suggests that Pros may function as a neuronal lineage-specific homeodomain protein cofactor involved in target selectivity during neuronal lineage development and thus may be the first example of a tissue-specific differential modulator of homeodomain proteins.
METHODS
Pros Protein Constructs.
L-Pros was created by cloning a 3.2-kb cDNA fragment encoding the C-terminal 1,000 amino acid residues of the Pros protein (residues 407-1406) into the pRSET expression vector. HxPros was constructed by PCR amplification of Pros DNA encoding the C-terminal 236 amino acid residues (residues 1171–1406) plus 32 base pairs of 3′ untranslated sequence, using the following primers: ATGGATCCATGCTCCATCCCGCCTTGCTG and GTAAGCTTCCTGCCGTTCGCCTTCGGATG. HxPros was cloned into the pRSET expression vector. ProsΔC was created by PCR deletion of the region C-terminal of Hx3 from HxPros, using the following primers: TTCGGTGACAGCTTGTCGTGC and CCATCCGAAGGCGAACGGCAG, which flank the deleted region and prime in opposite directions. The resulting PCR product was treated with DpnI to destroy methylated template molecules. Following extraction with phenol/chloroform and precipitation with ethanol, the PCR products were religated and used to transform Escherichia coli. ProsΔH/C represents a frameshift mutation of HxPros that replaced Hx3 by a heterologous sequence of 18 amino acids terminating in a stop codon. All constructs were sequenced to verify the introduced mutations.
Protein Expression.
Protein constructs in pRSET vector were used to transform E. coli DE21-LysE. After reaching an optical density at 600 nm of 0.3–0.4, cells were induced with 1 mM isopropyl β-d-thiogalactoside (IPTG) for 2–3 hr. After induction, proteins were harvested by collecting the insoluble fraction from lysed bacterial cells and solubilizing it in urea-5 buffer (20 mM Tris⋅HCl, pH 7.9/0.5 M NaCl/6 M urea/5 mM imidazole/0.1% Triton X-100) followed by sonication. The protein preparations were dialyzed against TCB buffer (20 mM Tris⋅⋅HCl, pH 8.0/150 mM KCl/2.5 mM CaCl2/1 mM phenylmethanesulfonyl fluoride/10 mM 2-mercaptoethanol) overnight at 4°C, divided into aliquots, and stored at −80°C. Dfd protein was prepared according to Zeng et al. (15). The Hoxa-5 PCR fragment was transcribed in vitro using the AmpliScribe T7 transcription system (Epicentre Technologies, Madison, WI) and translated by using rabbit reticulocyte lysate (Promega) according to specifications.
Protein Purification.
Solubilized pellets (see above) from L-Pros and HxPros proteins were loaded onto a column of 1 ml of Ni bead resin (Invitrogen). The column was washed with 10 ml of urea-5 and 4 ml of 10% urea-200/90% urea-5 (urea-200 is the same as urea-5 except that it contains 200 mM imidazole). The column was eluted with 4 ml of urea/EDTA (20 mM Tris⋅HCl, pH 7.9/0.5 M NaCl/6 M urea/100 mM EDTA). The eluted material was dialyzed against TCB buffer.
Target Detection Assay (TDA) and Electrophoretic Mobility-Shift Assay (EMSA).
TDA was based on a protocol by Thiesen and Bach (16) with the following modifications: Ni beads were washed twice with double-distilled H2O and twice with HE [25 mM potassium Hepes, pH 7.6/0.1 mM EDTA/0.1 mM dithiothreitol/10 mM KCl/10 mg/ml poly(dI)·poly(dC)]. HxPros and L-Pros proteins were immobilized on Ni beads and incubated with pools of double-stranded oligonucleotides with a random core of 12 base pairs and two flanking sequences used as primer annealing site in PCR (GTCGGATCCTGTCTGAGGTGAG-NNNNNNNNNNNN-GTCTTCCGACGTCGAATTCGCG). The primers used were GTCGGATCCTGTCTGAGGTGAG (primer A) and CGCGAATTCGACGTCGGAAGAC (primer B). PCR conditions were according to Thiessen and Bach (16). After incubation in HE at 4°C for 25 min, the beads were washed four times with 0.1 ml of HE, and bound DNA was eluted in 0.1 ml of double-distilled H2O, at 95°C for 5 min. Eluted DNA was amplified by PCR using primers complementary to the flanking sequences. After four such rounds of selection/amplification cycles, the DNA pools were used as probes in EMSA. Shifted material was excised, electroeluted, and reamplified. Two such EMSA/PCR rounds were performed. The reamplified material was inserted into Bluescript plasmid, and individual clones were sequenced. Probes for all EMSAs were end labeled with T4 polynucleotide kinase and incubated with bacterially expressed Pros proteins. Incubation was done in 1× KHEG (70–100 mM KCl/25 mM potassium Hepes, pH 7.6/0.1 mM EDTA/0.1 mM dithiothreitol/15 ng/μl salmon sperm DNA/10% glycerol] for 10 min on ice. The reaction mixtures were then loaded onto a nondenaturing 5% or 6% polyacrylamide/1× TBE gel and subjected to electrophoresis in 1× TBE (100 mM Tris/83 mM boric acid/1 mM EDTA, pH 8.3) at 4°C, 200–250 V. For interaction with homeodomain proteins, HxPros or ProsΔH/C was preincubated on ice with Dfd, Eve, or Hoxa-5 proteins for 2 min prior to addition of probes. The incubation was continued for an additional 8 min before loading on acrylamide gels as described above. The following double-stranded oligonucleotides were used in EMSA with HxPros (see Fig. 1 B, C, and D): TDA-Pros oligo (CTCATCGCTCTCATCGCTAG), ASE oligo (TTCGATGCATTTCTGCGTTTT0, and N-Box oligo (GTACGCCGGCACGCGACAGG) (17). EMSA gels were quantified by using PhosphorImager exposure cassettes and ImageQuant software (Molecular Dynamics).
Figure 1.
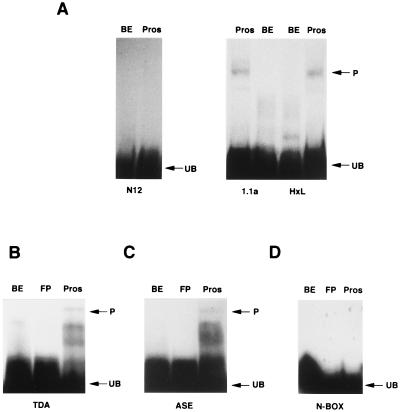
Pros binds specifically to a novel sequence. (A) TDA: HxPros binds to selected (1.1a, HxL) but not unselected (N12) pools of TDA oligonucleotides in EMSA. Oligonucleotides allowed to react with bacterial extracts (BE) lacking Pros protein are shown for control. (B–D) Pros binds to consensus oligonucleotides based on TDA-selected sequences (B) and to oligonucleotides based on a genomic sequence from the ase (T8) promoter (C) but not to a control N-Box oligonucleotide (D). P, position of Pros-bound material; FP, unreacted probe; UB, unbound probe.
Transfection Assays.
Transient transfection assays were performed in COS-1 cells. HxPros was expressed using the pCG expression vector (18). To obtain pML-mPbs-luc, a 60-bp oligonucleotide (CTTGGCTCAATACTAAGGATCTTGGCTCAATACTAAGGATCTTGGCTCAATACTAAGGAT) containing multiple Pros binding sites (mPbs) was cloned 5′ of the major late promoter driving the luciferase reporter gene (pML-Luc) (T. Hai, personal communication). Transfection of COS cells was performed as follows: 7 μg of total DNA in 0.25 M CaCl2/140 mM NaCl/5 mM KCl/0.75 mM Na2HPO4/6 mM dextrose/25 mM Hepes, pH 7.05, was used per 60-mm plate of 80% confluent COS-1 cells. Fourteen to 24 hr after transfection, cell media were changed, and cells were harvested in 250 mM Tris⋅HCl, pH 7.8, 24 hr after medium change. The cells were lysed by five freeze/thaw cycles and centrifuged for 2 min at 14,000 × g, and 0.1 ml of the supernatant was mixed with 0.1 ml of 1 mM luciferin (Sigma) and 0.3 ml of 25 mM Gly-Gly buffer, pH 7.8/5 mM ATP/15 mM MgSO4 and assayed for luciferase activity. All transfections were carried out in duplicate and all experiments were repeated at least three times.
Protein–Protein Interaction.
EMSA and interaction assays with Dfd were done as described above. Pros antibodies were added 5 min into the incubation with the probe. Incubation was continued for another 3 min. Trypsin proteolysis: 2 μg of purified Dfd protein was incubated with 5 μg of unpurified Pros protein or control bacterial lysate for 30 min on ice and then treated with 1 ng of trypsin for 1 min at 25°C. Proteolysis was stopped by adding an equal volume of 2× protein denaturation buffer and loaded on an SDS/12% polyacrylamide gel. After electrophoresis, proteins were transferred onto nitrocellulose filters and Dfd was detected by using polyclonal antibodies to Dfd (19). For Fig. 3C, 5 μg of Pros was incubated with 0.2, 1, and 2 μg of purified Dfd protein. Experiments were repeated at least three times.
Figure 3.
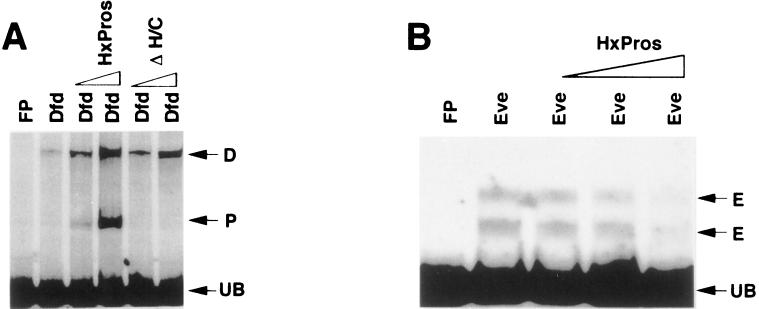
Pros differentially modulates the DNA-binding activity of homeodomain proteins. Pros enhances the DNA-binding activity of Dfd by 12-fold (A), while inhibiting Eve binding by 7-fold (B). The interaction between Pros and Dfd is independent of the DNA-binding activity of Pros, as ProsΔH/C can similarly enhance Dfd binding. P, D, and E, positions of probes bound by Pros, Dfd, and Eve, respectively. FP, unreacted probe; UB, unbound probe.
Immunohistochemistry.
Whole-mount wild-type and homozygous pros mutant embryos with one copy of the NAE600-LacZ construct were stained with anti β-galactosidase antibodies as described elsewhere (2). Genotypes were identified by using a blue balancer.
RESULTS
Pros Binds a Novel DNA Sequence.
To determine which DNA sequences are preferentially bound by Pros, two Pros protein constructs (L-Pros and HxPros) were used in a TDA (16) along with a 56-mer oligonucleotide containing a 12-bp random core. L-Pros includes amino acid residues 407-1406 (all amino acid positions are according to ref. 2) of the Pros protein, while HxPros encodes only the C-terminal 236 amino acid residues (residues 1171–1406). After four rounds of selection, pools of selected and unselected oligonucleotides were labeled and assayed for binding by HxPros in EMSAs. Pools selected independently by either L-Pros or HxPros were bound in these assays. In contrast, the unselected control oligonucleotide pools did not bind (Fig. 1A). Selected sequences aligned along a common 7-bp motif, from which a consensus sequence was determined (Table 1).
Table 1.
Consensus sequence bound by Pros
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequences selected in TDA | ||||||||||||||||
| t | C | A | T | c | g | C | T | a | t | c | g | |||||
| C | A | C | g | a | C | T | a | t | c | t | t | |||||
| g | t | g | g | a | C | A | C | c | t | C | T | |||||
| t | g | t | a | g | C | T | T | g | g | C | T | |||||
| t | t | c | c | t | C | A | T | c | t | A | C | |||||
| C | A | T | c | a | C | C | t | c | c | t | c | |||||
| c | g | a | a | t | C | A | C | g | t | C | T | |||||
| a | a | t | a | g | C | A | C | g | t | C | T | |||||
| a | t | g | t | c | C | A | A | t | a | C | T | |||||
| a | t | c | g | t | C | T | T | a | c | C | T | |||||
| c | t | g | t | a | C | A | T | g | a | C | C | |||||
| C | A | T | g | c | C | T | a | c | c | g | c | |||||
| C | A | C | t | g | C | T | a | c | g | a | t | |||||
| g | c | t | C | T | T | a | t | C | T | g | a | |||||
| g | a | t | g | t | C | A | T | t | g | C | T | |||||
| Number of bases found in TDA sequences at each consensus position | ||||||||||||||||
| A | 0 | 12 | 1 | 2 | 4 | 1 | 0 | |||||||||
| C | 15 | 0 | 5 | 4 | 2 | 14 | 3 | |||||||||
| G | 0 | 0 | 0 | 6 | 4 | 0 | 0 | |||||||||
| T | 0 | 3 | 9 | 3 | 5 | 0 | 12 | |||||||||
| Deduced consensus: | C | A/t | c/t | N | N | C | T/c | |||||||||
Uppercase letters indicate a strong preference.
The consensus sequence, C A/t c/t N N C T/c, showed strong preferences at five of the seven positions (positions 1, 2, 3, 6, and 7), while the other two positions (positions 4 and 5) appeared to be random. To test whether the 7-bp consensus sequence is sufficient for Pros binding, oligonucleotides containing tandem repeats of the consensus were generated. These oligonucleotides were bound by HxPros in EMSA (Fig. 1B). In contrast, control oligonucleotides were not bound (Fig. 1D). Next we asked if our consensus sequence is able to predict Pros binding sites in a known genetic target gene. For this a 21-bp fragment of the ase promoter containing a consensus Pros binding site was used with HxPros in EMSA. As expected, this fragment was bound by HxPros (Fig. 1C). It should be noted that the identified consensus Pros binding site differs from the typical homeodomain binding motif. While this may seem surprising, it must be emphasized that Pros is a highly divergent member of the homeodomain protein family. The difference in binding specificity of Pros may be one consequence of this divergence.
Pros Can Regulate Transcription by Means of Pros Binding Sites.
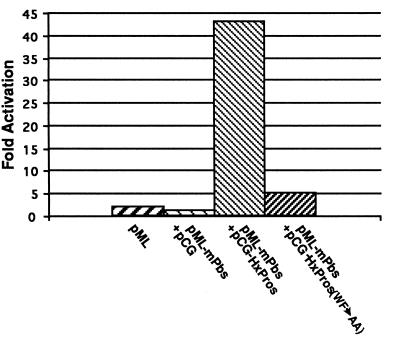
We then asked if Pros can mediate transcriptional regulation in a cell culture system by a 60-bp oligomer consisting of multiple Pros binding sites (mPbs) driving a luciferase reporter gene (pML-mPbs-luc). HxPros binds quite strongly to the mPbs oligo in vitro (not shown). In these experiments HxPros activated pML-mPbs-luc expression by an average of 43-fold over control transfections (Fig. 2). Thus in cell culture Pros can act as a transcription factor in the presence of Pros binding sites. To test whether DNA binding is required for HxPros-mediated transcription of the reporter gene construct, we tested the capacity of the Pros mutant HxProsWF-AA to activate expression of the pML-mPbs-luc reporter gene. In the HxProsWF-AA protein, the two highly conserved tryptophan and phenylalanine residues in the third helix of the Pros homeodomain were replaced with alanine residues. The resulting HxProsWF-AA protein does not bind to the Pros binding site in vitro (not shown). The loss of the capacity of HxProsWF-AA to bind DNA in vitro is paralleled by a strong reduction in the capacity of HxProsWF-AA to activate expression of the pML-mPbs-luc reporter gene (Fig. 2).
Figure 2.
Pros activates transcription by means of a 60-bp oligonucleotide consisting of multiple Pros binding sites (mPbs). HxPros activates the expression of a luciferase reporter gene in the presence of the mPbs oligo promoter region (pML-mPbs-luc + pCG-Pros). This activation is Pros dependent, as the expression vector (pCG) alone does not result in the activation of the reporter gene either in the absence (pML + pCG) or in the presence (pML-mPbs + pCG) of the mPbs oligo. Furthermore, the point mutant HxPros(WF→AA), which cannot bind Pros binding sites, shows a strongly reduced activation of the pML-mPbs-luc reporter gene.
Pros Modulates the DNA-Binding Properties of Homeodomain Proteins.
Given the panneural expression of Pros, its ability to act as a DNA-binding transcription factor may explain how it regulates panneural genes, but cannot easily explain how it can differentially regulate subset-specific expression of genes such as ftz, eve, en, and Dfd (see below). This raises the possibility that Pros cooperates with other transcription factors. Interestingly, the nervous system expression of eve and ftz each requires the other in addition to pros function (12, 20, 21). Therefore the proper expression of these homeobox genes in the nervous system appears to require the activities of both homeodomain proteins and Pros, suggesting that Pros may participate in homeodomain-dependent gene regulation during neuronal lineage development. To test whether Pros can interact with homeodomain proteins, we used two fragments (module-E, and N-100) from the regulatory regions of the Dfd gene, with typical homeodomain binding sites (15, 22) in close proximity to consensus Pros binding sites. N-100 is part of a 600-bp neuronal enhancer element of Dfd (N-600: see below) (22). We assayed the ability of Pros to modulate the DNA binding of different homeodomain proteins: the Drosophila Eve, En, and Dfd proteins, and the mouse Hoxa-5 protein (23). Homeodomain proteins play critical roles in the development of various cell lineages in many organisms, yet their in vitro binding sites are very similar (24). This has raised the question of how homeodomain proteins achieve differential tissue-specific gene regulation during development. It has been suggested that interactions with cofactors play a pivotal role in tissue-specific gene regulation (25, 26). In our experiments HxPros is capable of influencing the ability of some homeodomains to bind to DNA in vitro. HxPros enhances the binding of Dfd and Hoxa-5 to DNA by more than 10-fold (Fig. 3A, and not shown). Interestingly, this modulation varies with different homeodomain proteins. For example HxPros reduces Eve’s DNA binding to very low levels (Fig. 3B), whereas no modulation was observed with En (not shown). These in vitro results suggest that Pros may differentially influence the activity of different homeodomain proteins. Finally, this interaction is unidirectional and specific. Neither Dfd, Eve, nor En has an apparent effect on Pros binding (data not shown). Control bacterial lysates do not alter Dfd or Eve binding activity, and incubation with the homeodomain proteins Eve or En does not result in an enhancement of Dfd binding (data not shown; ref. 19).
The capacity of Pros to modulate the DNA-binding properties of homeodomain proteins in vitro does not require DNA binding of the Pros protein itself. A non-DNA-binding deletion construct of HxPros, ProsΔH/C (residues 1289–1406 deleted), was tested with Dfd and Hoxa-5. ProsΔH/C enhances the DNA-binding activity of both Dfd (Fig. 3B) and Hoxa-5 (not shown). Since ProsΔH/C maintains the capacity to modulate the DNA-binding activities of Dfd and Hoxa-5, the homeodomain protein interacting activity of HxPros can be separated from its DNA-binding activity, and it is localized in the 115 N-terminal amino acid residues of HxPros (residues 1171–1285). This region includes helices 1 and 2, but not helix 3 (Hx3), of the Pros homeodomain. This is consistent with the recent finding that residues in the N-terminal portion of the Pbx homeodomain, another atypical homeodomain protein, are required for protein-specific interactions with various homeodomain proteins (27).
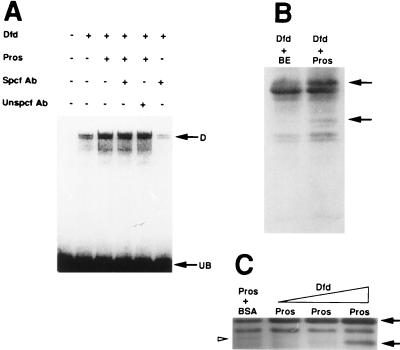
Under the conditions of our in vitro experiments, Pros does not appear to form heteromeric DNA-binding complexes with Dfd or Hoxa-5, as no increased retardation in the mobility of the Dfd–DNA or Hoxa-5–DNA complexes was apparent. We tested whether Pros is a part of the Dfd DNA-binding complex by adding polyclonal Pros antibodies to Dfd-ProsΔH/C EMSA experiments. We found that these antibodies, which supershifted a HxPros–DNA complex (data not shown), did not supershift a Pros-enhanced Dfd DNA-binding complex (Fig. 4A). This indicates that under the conditions of these experiments Pros is not a part of the Dfd–DNA complex, and further suggests that Pros interacts with Dfd to modify its DNA-binding capacity, but may not form a ternary DNA-binding complex with it. This type of interaction between Pros and Dfd is not similar to the interaction between Dfd and Exd (28). However, enhancement of DNA-binding activity in the absence of detectable ternary complexes has been reported for other homeodomain proteins in EMSA experiments (29, 30).
Figure 4.
(A) Pros is not detected in an enhanced Dfd DNA-binding complex. HxPros antibodies do not cause a retardation of the mobility of Pros-enhanced Dfd DNA binding to the Dfd neuronal element N-100. Symbols same as in Fig. 3. (B) Partial trypsin proteolysis of Dfd protein incubated with HxPros results in reproducible changes in the proteolytic pattern (arrows) as compared with the pattern generated when Dfd is incubated with control bacterial lysates. (C) Increasing concentrations of Dfd cause changes in the proteolytic pattern of HxPros (arrows, open arrowhead) as compared with the pattern generated when HxPros is incubated with bovine serum albumin.
One possible mechanism for the modulation by Pros of Dfd DNA-binding activity could involve Pros-mediated conformational changes, which may expose domains essential for higher affinity binding, as seen between Labial and Exd (31). To test this possibility, Dfd protein was allowed to react with either HxPros or control bacterial lysate and subjected to partial trypsin proteolysis (32). Dfd protein reacted with HxPros shows at least two distinct and reproducible changes in the band pattern generated by partial trypsin proteolysis as compared with the pattern generated when Dfd is incubated with control lysate (Fig. 4B), suggesting a conformational change in Dfd upon interaction with Pros. Parallel conformational changes are seen in HxPros upon interaction with Dfd (Fig. 4C). These interactions do not require the presence of DNA.
Pros Is Required for Proper Regulation of a Dfd-Dependent Neural Enhancer.
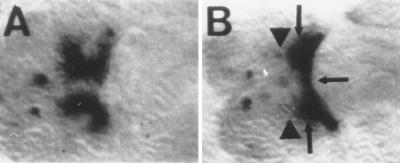
To test whether the interaction of Pros with Dfd may have consequences on the function of Dfd during nervous system development, we examined the expression pattern of a lacZ reporter gene under the control of a neuronal autoregulatory enhancer of Dfd (N-600) which drives LacZ expression in the subesophageal ganglion in a Dfd-dependent manner (22). Pros binds to this element and enhances Dfd binding to it (Fig. 4A) In pros mutant embryos LacZ expression is missing in a subset of cells of the subesophageal ganglion (Fig. 5B) as compared with LacZ expression in wild-type embryos (Fig. 5A). In addition, ectopic expression of the reporter gene is evident in a subset of subesophageal cells in pros mutant embryos (Fig. 5B). These data suggest that Pros is required along with Dfd for proper transcriptional regulation mediated by the neuronal enhancer, most likely by modifying the DNA-binding capacity of Dfd.
Figure 5.
Pros regulates a neural autoregulatory element of Dfd. Blow-up of the subesophageal region of stage 15 wild-type (A) and homozygous pros mutant (B) embryos. (A) An intronic enhancer element of Dfd drives the expression of LacZ in the subesophageal ganglion in the central nervous system. (B) Elements of LacZ expression are missing in stage 15 homozygous pros mutant embryos (arrowheads), while ectopic expression appears in other subsets of subesophageal cells (arrows). (×1,000.)
DISCUSSION
The panneural neuronal precursor gene pros belongs to a class of genes postulated to act early during neuronal lineage development and differentiation (reviewed in ref. 33). In the developing central and peripheral nervous systems, Pros is expressed in the nuclei of secondary precursors that will divide to give rise to neurons and glial cells, where Pros is also localized to the nuclei. In pros mutations the secondary precursors, as well as their progeny, exhibit aberrant gene expression. Thus the onset of the molecular mutant phenotype of Pros parallels its nuclear localization, indicating that Pros must be present in the nucleus for it to function. Consistent with this model, we have shown that Pros is a novel sequence-specific DNA-binding protein and acts as a transcription factor.
Homologues of Pros have been identified in vertebrate and invertebrate systems. The homology between these proteins is mainly limited to the C-terminal region, which is sufficient for DNA binding, transcriptional activity, and interaction with homeodomain proteins. If, as in typical homeodomain proteins, helix 3 (Hx3) of the Pros homeodomain also mediates DNA binding, its absolute conservation in all Pros homologues indicates that these proteins may recognize similar binding sites. In addition, the binding site of the Drosophila Pros protein is different from the binding sites of typical homeodomain proteins and other identified transcription factors. Thus, Pros defines a novel subclass of evolutionarily conserved transcription factors. In both mouse and chicken, the pros RNA expression pattern suggests a role in neuronal lineage development (7, 9). In addition, the ability of Drosophila Pros to interact with the mouse Hoxa-5 protein suggests that this function is conserved in the vertebrate homologues of Pros.
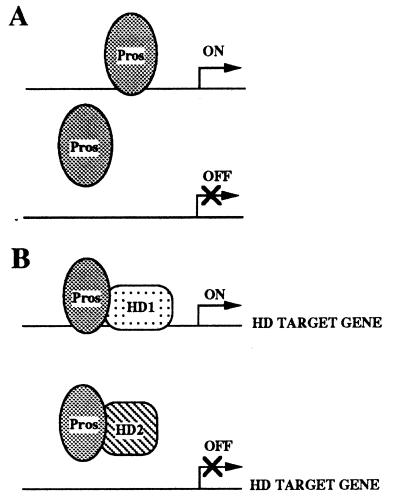
The diversity of aberrant gene expression and nervous system defects associated with pros mutations suggests that Pros may play multiple differential roles during neuronal lineage development (1–3). pros mutations result in morphological defects in multiple types of neurons in most or all neuronal lineages (2, 12) as well as in differentiation defects in glial cells (14). At different stages, and in different lineages, some Pros target genes are repressed, whereas others are ectopically expressed, indicating that Pros function is required for activation and repression, depending on the specific cellular environment. Given that many homeodomain proteins bind to rather similar core DNA sequences (24, 34, 35), it has been suggested that homeodomain proteins obtain additional specificity through interactions with cofactors. For example, the Extradenticle (Exd) protein is required for transcriptional regulation of target genes by homeotic selector proteins (36). In addition, both Exd and its vertebrate homologues, the PBX family of proteins, can modulate the DNA-binding activity of several homeodomain proteins (29, 37–40). However, while Exd has been shown to enhance the DNA-binding activity of homeodomain proteins, the rather ubiquitous expression of Exd does not easily explain a role in differential tissue-specific gene regulation by homeodomain proteins. The ability of Pros to either enhance or inhibit the DNA-binding properties of homeodomain proteins, as well as its neuronal lineage-specific expression, suggest that it may represent a neural-specific homeodomain protein cofactor. The observation that Pros activity, in addition to the activity of the homeodomain protein Dfd, is required for the proper expression of the Dfd neural autoregulatory element supports this idea. This presents a possible model to explain the diverse requirements for Pros in gene regulation. In this model Pros could function by binding to regulatory regions and directing activation or repression (Fig. 6A). Pros could also differentially enhance or inhibit the association of various homeodomain proteins with regulatory regions of their genes (Fig. 6B). This would allow Pros to contribute to the temporal and spatial specificity of gene regulation by homeodomain proteins during neuronal lineage development.
Figure 6.
Model for Pros function. (A) Pros may function as an independent transcription factor whose regulatory activity is mediated by direct binding to its target sites. (B) Pros may also function as a cofactor for various homeodomain proteins (HD) to differentially modulate their ability to bind to similar target sites. This differential modulation would result in the preferential binding of one homeodomain protein to a specific site, while at the same time inhibiting the binding of another homeodomain protein to the same site.
Acknowledgments
We are grateful to Drs. W. McGinnis, T. Hai, W. Herr, and J. Oberdick for providing constructs for this study. We thank Drs. Hai and Oberdick, as well as Mauricio Ramirez, James Smith, Mary Weiler, and the members of the Vaessin laboratory for helpful discussions. The research was supported by grants from the National Science Foundation and the Alfred P. Sloan Foundation, and by a seed grant from Ohio State University to H.V.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TDA, target detection assay; EMSA, electrophoretic mobility-shift assay.
References
- 1.Chu-LaGraff, Q., Wright, D. M., McNeil, L. K. & Doe, C. Q. (1991) Development Suppl, 79–85. [PubMed]
- 2.Vaessin H, Grell E, Wolf E, Bier E, Jan L Y, Jan Y N. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 3.Matsuzaki F, Koizumi K, Hama C, Yoshioka T, Nabeshima Y-I. Biochem Biophys Res Commun. 1992;182:1326–1332. doi: 10.1016/0006-291x(92)91878-t. [DOI] [PubMed] [Google Scholar]
- 4.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Nature (London) 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 5.Knoblich J A, Jan L Y, Jan Y N. Nature (London) 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 6.Spana E P, Doe C Q. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- 7.Oliver G, Sosa-Pineda B, Geisendorf S, Spana E P, Doe C Q, Gruss P. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- 8.Bürglin T R. Trends Biochem Sci. 1994;19:70–71. doi: 10.1016/0968-0004(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 9.Tomarev S I, Sundin O, Banerjee-Basu S, Duncan M K, Yang J-M, Piatigorsky J. Dev Dyn. 1996;206:354–367. doi: 10.1002/(SICI)1097-0177(199608)206:4<354::AID-AJA2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 10.Zinovieva R D, Duncan M K, Johnson T R, Torres R, Polymeropoulos M H, Tomarev S I. Genomics. 1996;35:517–522. doi: 10.1006/geno.1996.0392. [DOI] [PubMed] [Google Scholar]
- 11.Bürglin T R. In: Guidebook to the Homeobox Genes. Duboule D, editor. Oxford: Oxford Univ. Press; 1994. pp. 25–72. [Google Scholar]
- 12.Doe C Q, Chu-LaGraff Q, Wright D M, Scott M P. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- 13.Broadie K, Bate M. Nature (London) 1993;361:350–353. doi: 10.1038/361350a0. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs J R. J Neurobiol. 1993;24:611–626. doi: 10.1002/neu.480240507. [DOI] [PubMed] [Google Scholar]
- 15.Zeng C, Pinsonneault J, Gellon G, McGinnis N, McGinnis W. EMBO J. 1994;13:2362–2377. doi: 10.1002/j.1460-2075.1994.tb06520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiesen H-J, Bach C. Nucleic Acids Res. 1990;18:3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Herr W. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 19.Pinsonneault J. Ph.D. thesis. New Haven, CT: Yale Univ.; 1997. [Google Scholar]
- 20.Doe C Q, Smouse D, Goodman C S. Nature (London) 1988;333:376–378. doi: 10.1038/333376a0. [DOI] [PubMed] [Google Scholar]
- 21.Doe C Q, Hiromi Y, Gehring W J, Goodman C S. Science. 1988;239:170–175. doi: 10.1126/science.2892267. [DOI] [PubMed] [Google Scholar]
- 22.Lou L, Bergson C, McGinnis W. Nucleic Acids Res. 1995;23:3481–3487. doi: 10.1093/nar/23.17.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colberg-Poley A M, Voss S D, Chowdhury K, Gruss P. Nature (London) 1985;314:713–718. doi: 10.1038/314713a0. [DOI] [PubMed] [Google Scholar]
- 24.Desplan C, Theis J, O’Farrel P. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick A, Core N, Kerridge S, Scott M P. Development. 1995;121:2799–2812. doi: 10.1242/dev.121.9.2799. [DOI] [PubMed] [Google Scholar]
- 26.Wilson D S, Desplan C. Curr Biol. 1995;5:32–34. doi: 10.1016/s0960-9822(95)00010-8. [DOI] [PubMed] [Google Scholar]
- 27.Peltenburg L T C, Murre C. Development. 1997;124:1089–1098. doi: 10.1242/dev.124.5.1089. [DOI] [PubMed] [Google Scholar]
- 28.Pinsonneault J, Florence B, Vaessin H, McGinnis W. EMBO J. 1997;16:2032–2042. doi: 10.1093/emboj/16.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grueneberg D A, Natesan S, Alexandre C, Gilman M Z. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 30.Chan S K, Jaffe L, Capovilla M, Botas J, Mann R S. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 31.Chan S K, Mann R S. Proc Natl Acad Sci USA. 1996;93:5223–5228. doi: 10.1073/pnas.93.11.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan S K, Popperl H, Krumlauf R, Mann R S. EMBO J. 1996;15:2476–2487. [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan B, Vaessin H. Dev Genet. 1996;18:18–27. doi: 10.1002/(SICI)1520-6408(1996)18:1<18::AID-DVG3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Desplan C, Theis J, O’Farrel P. Nature (London) 1985;318:630–635. doi: 10.1038/318630a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoey T, Levine M. Nature (London) 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- 36.Rauskolb C, Wieschaus E. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Dijk M A, Murre C. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 38.Chang C-P, Shen W-F, Rozenfeld S, Lawrence H J, Largman C, Cleary M. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 39.Phelan M L, Rambaldi I, Featherstone M S. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Dijk M A, Peltenburg L T, Murre C. Mech Dev. 1995;52:99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]