Abstract
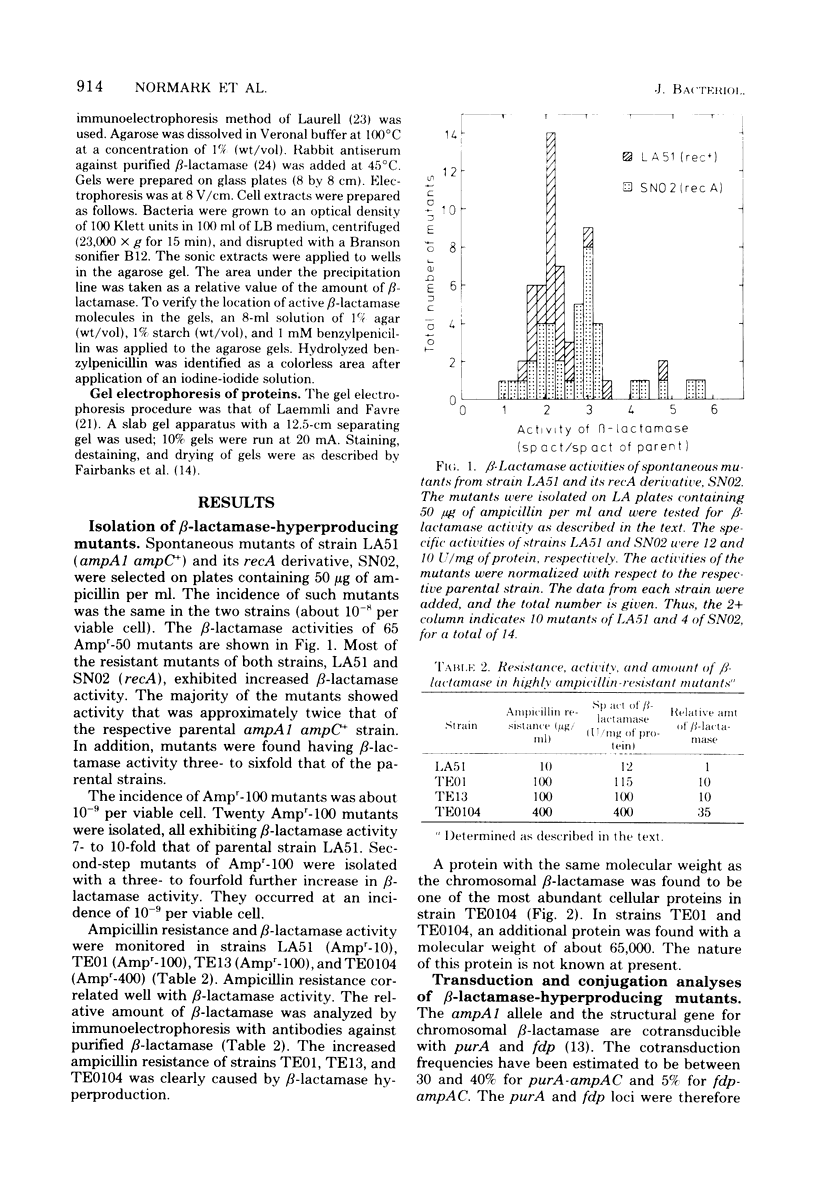
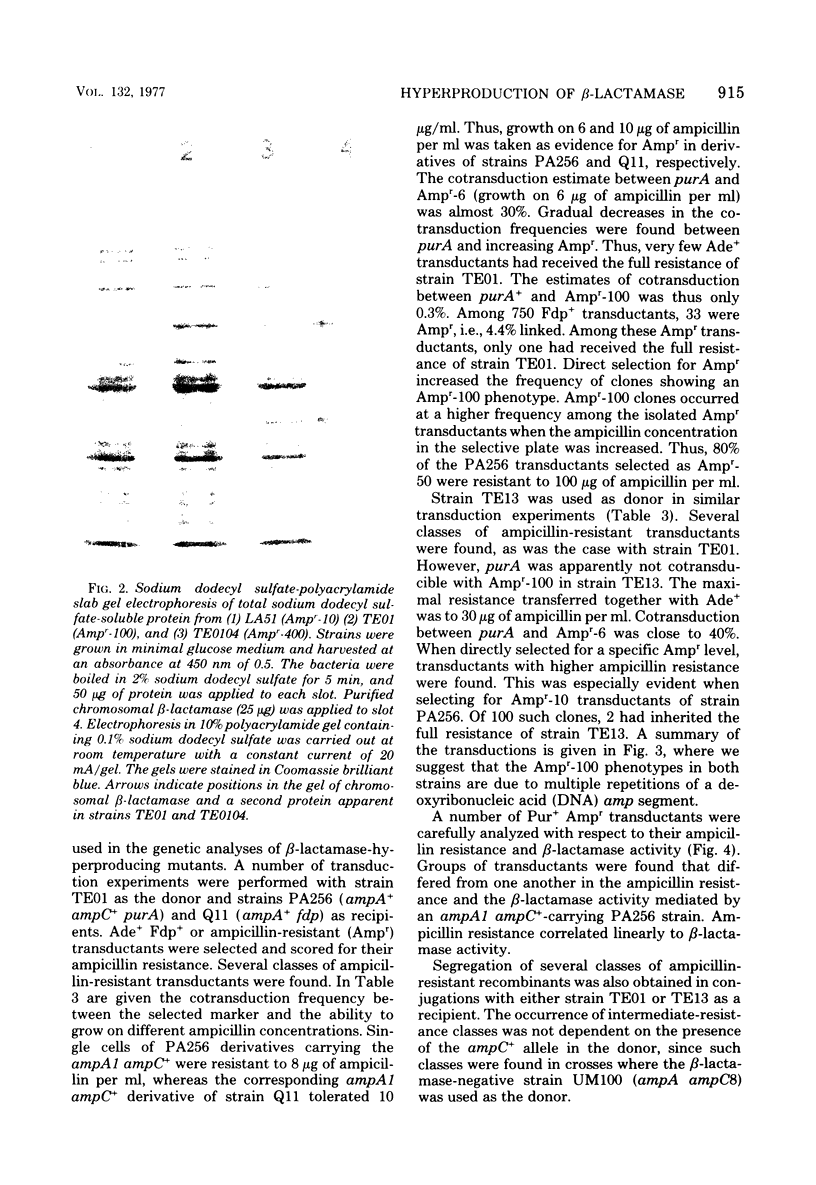
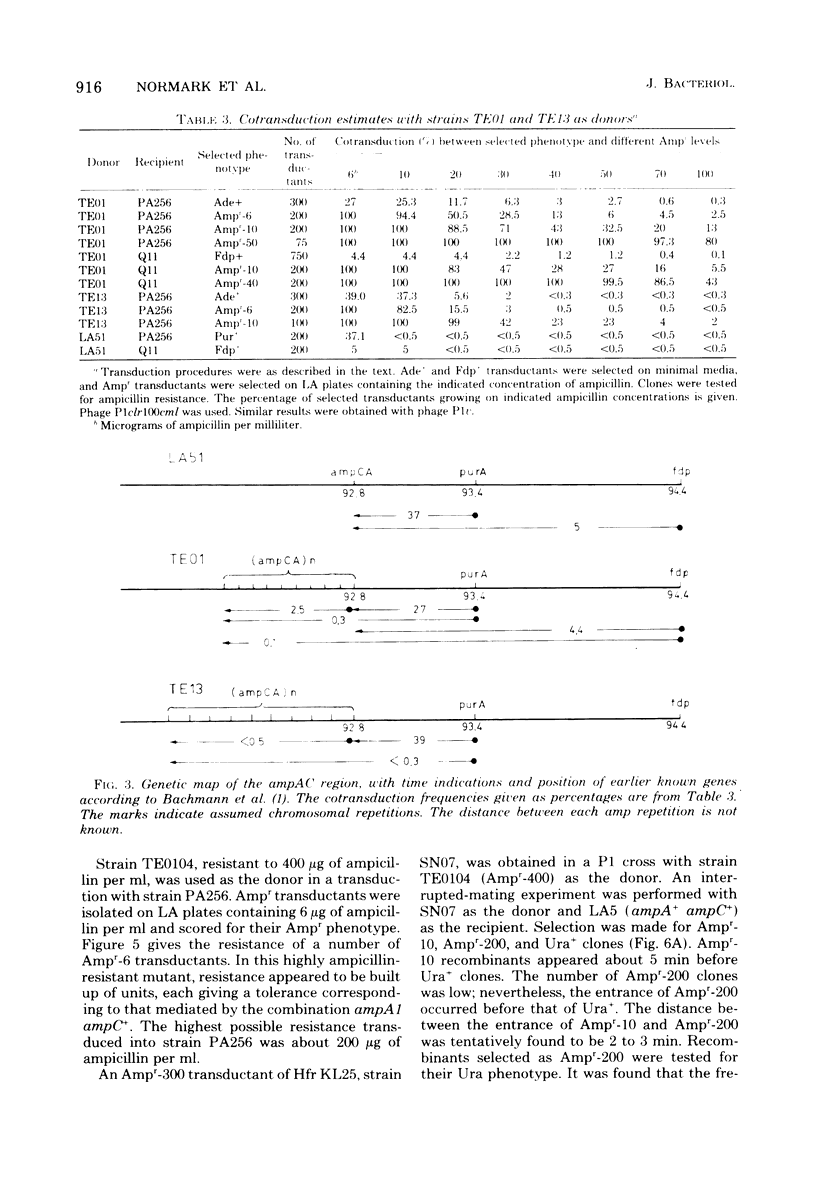
Escherichia coli K-12 ampicillin-resistant mutants hyperproducing chromosomal β-lactamase arose spontaneously from strains carrying ampA1 ampC+. Such mutants were found even in a recA background. Two Ampr-100 strains were analyzed genetically. The Ampr-100 resistance level of both strains could be transduced by direct selection for ampicillin resistance. Several classes of ampicillin-resistant transductants were found that differed from one another in the β-lactamase activity and the ampicillin resistance mediated by an ampA1 ampC+-carrying strain. The data suggested that β-lactamase hyperproduction was due to repetitions of the chromosomal amp genes. The size of the repeated region was calculated from cotransduction estimates, using the formula of Wu (Genetics 54:405-410, 1966), and was found to be about 1 min in one strain and 1.5 min in the other. Second-step Ampr-400 mutants were isolated from an Ampr-100 strain. The resistance of these mutants was apparently also due to repetitions, each mediating a resistance to about 10 μg/ml. Mutants of wild-type strains that were moderately resistant to ampicillin also gave rise to intermediate-resistance classes, suggesting repetitions of the wild-type amp alleles. F′ factors hyperproducing chromosomal β-lactamase by gene repetitions were constructed. They mediated levels of ampicillin resistance comparable to that of naturally occurring resistance plasmids. The expression of β-lactamase hyperproduction was not affected by the presence of ampA and ampC alleles in trans and did not act in trans on the other alleles.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeftinck F., Cunin R., Glansdorff N. Arginine gene duplications in recombination proficient and deficient strains of Escherichia coli K 12. Mol Gen Genet. 1974;132(3):241–253. doi: 10.1007/BF00269397. [DOI] [PubMed] [Google Scholar]
- Bennett P. M., Richmond M. H. Translocation of a discrete piece of deoxyribonucleic acid carrying an amp gene between replicons in Eschericha coli. J Bacteriol. 1976 Apr;126(1):1–6. doi: 10.1128/jb.126.1.1-6.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski M. M., Matthew M., Barth P. T., Datta N., Grinter N. J., Jacob A. E., Kontomichalou P., Dale J. W., Smith J. T. Plasmid-determined beta-lactamase indistinguishable from the chromosomal beta-lactamase of Escherichia coli. J Bacteriol. 1976 Jan;125(1):149–157. doi: 10.1128/jb.125.1.149-157.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman H. G., Monner D. A. Characterization of lipopolysaccharides from Escherichia coli K-12 mutants. J Bacteriol. 1975 Feb;121(2):455–464. doi: 10.1128/jb.121.2.455-464.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman H. G., Nordström K., Normark S. Penicillin resistance in Escherichia coli K12: synergism between penicillinases and a barrier in the outer part of the envelope. Ann N Y Acad Sci. 1974 May 10;235(0):569–586. doi: 10.1111/j.1749-6632.1974.tb43291.x. [DOI] [PubMed] [Google Scholar]
- Burman L. G., Nordström K. Colicin tolerance induced by ampicillin or mutation to ampicillin resistance in a strain of Escherichia coli K-12. J Bacteriol. 1971 Apr;106(1):1–13. doi: 10.1128/jb.106.1.1-13.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman L. G., Park J. T., Lindström E. B., Boman H. G. Resistance of Escherichia coli to penicillins: identification of the structural gene for the chromosomal penicillinase. J Bacteriol. 1973 Oct;116(1):123–130. doi: 10.1128/jb.116.1.123-130.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdá-Olmedo E., Hanawalt P. C., Guerola N. Mutagenesis of the replication point by nitrosoguanidine: map and pattern of replication of the Escherichia coli chromosome. J Mol Biol. 1968 May 14;33(3):705–719. doi: 10.1016/0022-2836(68)90315-x. [DOI] [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Eriksson-Grennberg K. G. Resistance of Escherichia coli to penicillins. II. An improved mapping of the ampA gene. Genet Res. 1968 Oct;12(2):147–156. doi: 10.1017/s0016672300011769. [DOI] [PubMed] [Google Scholar]
- Fairbanks G., Steck T. L., Wallach D. F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971 Jun 22;10(13):2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- HORIUCHI T., HORIUCHI S., NOVICK A. The genetic basis of hyper-synthesis of beta-galactosidase. Genetics. 1963 Feb;48:157–169. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Combriato G. Genetic duplications induced at very high frequency by ultraviolet irradiation in Escherichia coli. Mol Gen Genet. 1973 Dec 31;127(3):197–214. doi: 10.1007/BF00333760. [DOI] [PubMed] [Google Scholar]
- Hu S., Ohtsubo E., Davidson N. Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. J Bacteriol. 1975 May;122(2):749–763. doi: 10.1128/jb.122.2.749-763.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko D. J., Cohen S. N. Site specific recA--independent recombination between bacterial plasmids: involvement of palindromes at the recombinational loci. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1373–1377. doi: 10.1073/pnas.72.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Langridge J. Mutations conferring quantitative and qualitative increases in beta-galactosidase activity in Escherichia coli. Mol Gen Genet. 1969;105(1):74–83. doi: 10.1007/BF00750315. [DOI] [PubMed] [Google Scholar]
- Laurell C. B. Quantitative estimation of proteins by electrophoresis in agarose gel containing antibodies. Anal Biochem. 1966 Apr;15(1):45–52. doi: 10.1016/0003-2697(66)90246-6. [DOI] [PubMed] [Google Scholar]
- Linström E. B., Boman H. G., Steele B. B. Resistance of Escherichia coli to penicillins. VI. Purification and characterization of the chromosomally mediated penicillinase present in ampA-containing strains. J Bacteriol. 1970 Jan;101(1):218–231. doi: 10.1128/jb.101.1.218-231.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monner D. A., Jonsson S., Boman H. G. Ampicillin-resistant mutants of Escherichia coli K-12 with lipopolysaccharide alterations affecting mating ability and susceptibility to sex-specific bacteriophages. J Bacteriol. 1971 Aug;107(2):420–432. doi: 10.1128/jb.107.2.420-432.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Boman H. G., Matsson E. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol. 1969 Mar;97(3):1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Burman L. G. Resistance of Escherichia coli to penicillins: fine-structure mapping and dominance of chromosomal beta-lactamase mutations. J Bacteriol. 1977 Oct;132(1):1–7. doi: 10.1128/jb.132.1.1-7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odakura Y., Tanaka T., Hashimoto H., Mitsuhashi S. Mutation of R factors capable of specifying hypersynthesis of penicillinase. Antimicrob Agents Chemother. 1973 Mar;3(3):315–324. doi: 10.1128/aac.3.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Stickgold R. Selective amplification of genes on the R plasmid, NR1, in Proteus mirabilis: an example of the induction of selective gene amplification. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2518–2522. doi: 10.1073/pnas.74.6.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]