Abstract
The Drosophila retinal degeneration C (rdgC) gene encodes an unusual protein serine/threonine phosphatase in that it contains at least two EF-hand motifs at its carboxy terminus. By a combination of large-scale sequencing of human retina cDNA clones and searches of expressed sequence tag and genomic DNA databases, we have identified two sequences in mammals [Protein Phosphatase with EF-hands-1 and 2 (PPEF-1 and PPEF-2)] and one in Caenorhabditis elegans (PPEF) that closely resemble rdgC. In the adult, PPEF-2 is expressed specifically in retinal rod photoreceptors and the pineal. In the retina, several isoforms of PPEF-2 are predicted to arise from differential splicing. The isoform that most closely resembles rdgC is localized to rod inner segments. Together with the recently described localization of PPEF-1 transcripts to primary somatosensory neurons and inner ear cells in the developing mouse, these data suggest that the PPEF family of protein serine/threonine phosphatases plays a specific and conserved role in diverse sensory neurons.
Light detection by both vertebrate and invertebrate retinal photoreceptors proceeds through the activation of visual pigments and their G protein targets. However, subsequent steps in the phototransduction cascades differ. Vertebrate phototransduction involves activation of a cGMP phosphodiesterase, hydrolysis of cGMP, and closure of cGMP-gated cation channels in the plasma membrane (1). Channel closure produces a hyperpolarization and a decrease in cytosolic calcium, which activates a feedback loop responsible for light adaptation (2). Invertebrate phototransduction, although less defined, appears to involve the activation of phospholipase C with a resulting increase in cytosolic calcium (3). Genetic analyses in Drosophila have revealed a high degree of structural and functional similarity between invertebrate and vertebrate systems in those aspects of phototransduction that relate directly to visual pigment function. In addition to the similarity with respect to G protein coupling noted above, both classes of photoreceptors turn off photoactivated visual pigment by phosphorylation and binding by a soluble cytosolic protein, arrestin (4, 5).
Similarities between vertebrates and invertebrates are also apparent in the deleterious effects of mutation on photoreceptor function and viability (6, 7). In the most extensively studied example, a close resemblance has been found between mutations in the Drosophila R1–R7 rhodopsin gene that cause retinal degeneration and mutations in the human rhodopsin gene that cause autosomal dominant retinitis pigmentosa (8–11). In both species, a variety of single amino acid substitutions perturb protein stability, folding, or transportation and produce an autosomal dominant phenotype. A resemblance is also apparent in arrestin gene mutations, which, in both species lead to prolonged activation of rhodopsin, producing in humans a characteristic elevation of the rod threshold and nightblindness (Oguchi disease; refs. 12 and 13). These similarities suggest that identifying human homologues of Drosophila phototransduction genes may be an efficient way to isolate candidate genes for human retinal diseases.
The retinal degeneration produced in Drosophila by loss of retinal degeneration C (rdgC) function requires both normal levels of rhodopsin and light exposure (10, 14). Drosophila rdgC is a divergent member of the PPP family of protein serine/threonine phosphatases and is unusual in possessing a large carboxy-terminal domain with at least two EF-hand motifs (15). Rhodopsin dephosphorylation in cellular homogenates and in vivo is abolished by mutations in rdgC, indicating that rdgC acts either directly or indirectly to effect rhodopsin dephosphorylation (5, 16). Curiously, the degeneration produced by mutation of rdgC is not suppressed by loss-of-function mutations in norpA (14) or in Dgq (16), the genes encoding the photoreceptor-specific phospholipase C and G protein α-subunit, respectively. These data suggest either that rdgC dephosphorylates substrates other than rhodopsin that are relevant to the retinal degeneration phenotype, or that a pathway distinct from that which mediates phototransduction is involved in the degeneration phenotype.
Here we report the identification and characterization of two rdgC homologues from mammals and one from C. elegans. In mammals, one of the homologues is expressed specifically in retinal photoreceptors and pinealocytes. Together with Drosophila rdgC, these new sequences define a distinctive and highly conserved branch of the protein serine/threonine phosphatase family.
MATERIALS AND METHODS
Large-Scale cDNA Sequencing.
Details of retina cDNA sequence determination are reported in ref. 17.
cDNA and Genomic Sequences.
The complete coding region sequences of human and mouse PPEF-2 each were obtained from two or more independent clones isolated by DNA hybridization to oligo(dT)- primed cDNA libraries from adult human retinas (18) and P0–P7 mouse eyes (A. Lanahan, H.S., and J.N., unpublished data), respectively. To isolate the human PPEF-2 gene, a Sau3AI partial-digest human genomic library in lambda EMBL3 was screened with full-length human PPEF-2 cDNA. The coding region sequence of PPEF-1 was obtained from two partial-length human retina cDNA clones and from an uncloned PCR product generated by 5′ RACE (rapid amplification of cDNA ends; ref. 19) using human fetal brain cDNA (CLONTECH) as template. The sequence of the 5′ proximal 1.3 kb of the C. elegans PPEF transcript was obtained from an uncloned PCR product generated by 5′ RACE using as template cDNA prepared from larval mRNA. PCR amplification was performed with gene-specific primers and an SL-1 spliced leader primer (20).
Northern Blot Hybridization.
RNA was isolated from adult rat tissues using the guanidinium thiocyanate method (21), and 20 μg of total RNA from each tissue was resolved by agarose gel electrophoresis in the presence of formaldehyde. The filter was hybridized with a 1-kb human PPEF-2 probe encompassing exons 1–9 in 50% formamide, 5× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA (SSPE) at 42°C, and washed in 0.2× SSPE at 50°C.
In Situ Hybridization.
In situ hybridization was performed with digoxigenin-labeled riboprobes (22) and 20-μm sections cut from unfixed albino rat eyes and brain frozen in OCT or from macaque retinas that had been perfused with PBS, 4% paraformaldehyde, cryoprotected in 30% sucrose, and frozen in OCT. An extra incubation of 30 min in PBS, 1% Triton-X 100 was applied to macaque retina sections immediately after the acetylation step. For hybridization to macaque sections, the riboprobe encompassed the first 492 codons of human PPEF-2. For hybridization to rat and mouse sections, the riboprobe encompassed codons 63–454 of mouse PPEF-2. Control riboprobes were synthesized from a macaque rhodopsin cDNA segment encoding amino acids 133–254 and a full-length mouse blue cone pigment cDNA (23).
Antibody Production and Purification.
Bacteriophage T7 gene 10 fusion proteins containing at their carboxy termini amino acids 542–753 from human PPEF-2(L) or 542–598 from human PPEF-2(S) were produced in Escherichia coli, purified by preparative SDS/PAGE, and used for immunization of rabbits (24). A corresponding pair of maltose-binding protein (MBP) fusions (New England Biolabs) containing the same PPEF-2 segments were produced in E. coli and purified to apparent homogeneity by amylose-affinity chromatography. The purified MBP fusions were used for affinity-purification of immune sera after being covalently coupled to Affi-Gel-15 (Bio-Rad).
Immunoblotting.
Rod outer segments were prepared by sucrose gradient centrifugation from dark-adapted bovine retinas as described (25). Bovine retinas or rod outer segments were resuspended in PBS, 0.5% Triton-X 100 containing protease inhibitors (1 μg/ml chymostatin, leupeptin, antipain, and pepstatin A; 2 μg/ml aprotinin; and 100 μg/ml phenylmethylsulfonyl fluoride), incubated on ice for 15 min, and spun at 5,000 g for 10 min to remove nuclei and debris. Protein concentrations were determined using the Bradford assay (Bio-Rad). Immunoblots were sequentially incubated with affinity-purified rabbit anti-human PPEF-2 antibodies and horseradish peroxidase-conjugated goat anti-rabbit antibody, and the antibodies were visualized using the ECL detection system (Amersham).
Immunohistochemistry.
Macaque retinas were harvested following cardiac perfusion with PBS, 4% paraformaldehyde, and overnight fixation of isolated eye cups in the same buffer. After cryoprotection in 8% sucrose and embedding in OCT, 10-μm frozen sections were prepared and preincubated for 1 hr in PBS containing 5% normal goat serum and 0.3% Triton-X 100 and then incubated in the same buffer containing the primary antibody overnight at 4°C. Biotinylated goat anti-rabbit secondary antibody and Texas Red-conjugated streptavidin (Vector Laboratories) or avidin-horseradish peroxidase (Extravidin-peroxidase, Sigma) were used to visualize PPEF-2 immunostaining. The cGMP-gated channel was visualized with mouse monoclonal PMc1D1 (26) and fluorescein-conjugated goat anti-mouse secondary antibody. Sections were viewed using a Noran Instruments confocal microscope (Middleton, WI). Bovine retinas were processed similarly except that fresh eye-cups were fixed at 4°C in PBS/4% paraformaldehyde for 3 hr, cryoprotected in 30% sucrose overnight, and embedded in OCT.
RESULTS
Identification and Characterization of PPEF-2.
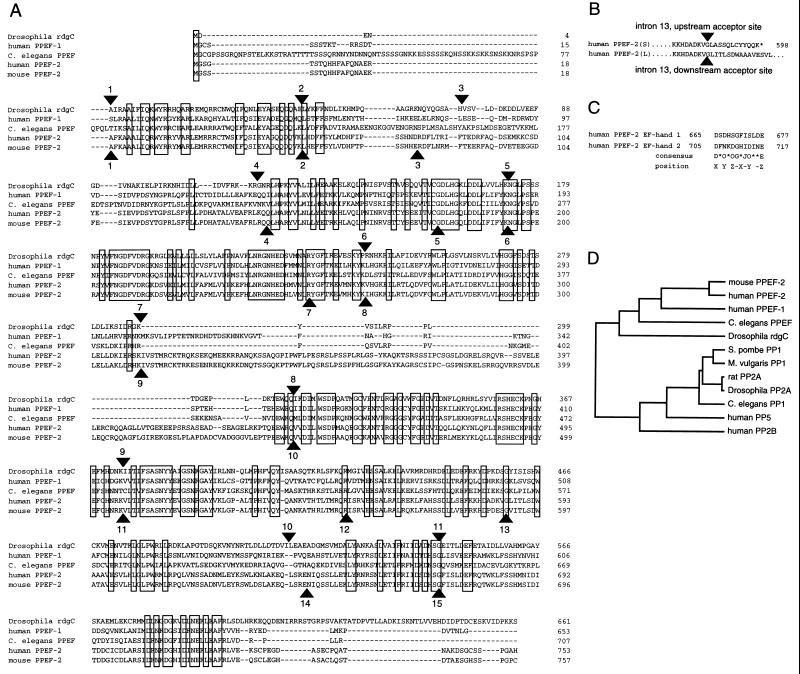
To efficiently identify novel genes involved in human retinal function and disease, we have determined partial sequences from 5,000 human retina cDNA clones (J.P.M., P.M.S., J.W., A. Rattner, and J.N., unpublished data). Conceptual translation of one of these sequences showed significant homology to the carboxy-terminal third of the Drosophila rdgC protein. This rdgC-like sequence, henceforth referred to as PPEF-2 (Protein Phosphatase with EF-hands-2), was of interest because it represented the first rdgC homologue in a species other than Drosophila. Isolation of cDNA clones from human retina and mouse eye libraries and characterization of their complete coding sequences revealed ORFs of 753 and 757 aa, respectively (Fig. 1). In the adult human retina cDNA library, PPEF-2 clones are found at a frequency of approximately 0.05%. The human and murine proteins show 82% mutual identity and 39% identity with Drosophila rdgC. Like rdgC, the mammalian homologues possess two regions near the carboxy terminus that closely match the EF-hand consensus for predicted calcium-liganding residues (Fig. 1C; ref. 27). One additional region of poorer homology is found between amino acids 581 and 592 (in the human PPEF-2 numbering system). Significant homology to the more distantly related members of the PPP family of protein serine/threonine phosphatases is present in the catalytic domain, which is predicted to reside between amino acids 135 and 535 in human PPEF-2 (28–30).
Figure 1.
(A) Alignment of amino acid sequences from human, mouse, D. melanogaster, and C. elegans PPEF family members. An Arg/Ser polymorphism was found at position 118 in human PPEF-2. Arrowheads beneath the sequences indicate intron positions in human PPEF-2; arrowheads above the sequences indicate intron positions in Drosophila rdgC. Residues that are identical in all five sequences are boxed. The alignment was performed using geneworks software. (B) Differential splicing to either of two acceptor sites in exon 14 generates short or long versions of PPEF-2. The point of divergence is indicated by the arrowheads. The asterisk indicates a stop codon. (C) Comparison of the calcium chelation loop of two human PPEF-2 EF-hands with the EF-hand consensus sequence (27). O, amino acid with an oxygen atom in its side-chain (D, N, E, Q, S, or T); J, (I, L, or V); ∗, any amino acid. (D) Sequence similarity between PPEF family members and a representative subset of PPP phosphatases. The lengths of the horizontal lines in the dendrogram are proportional to the degree of amino acid divergence. C. elegans PPEF and the C. elegans PP1 homologue represent the two C. elegans sequences that are most similar to the PPEF family. The sequences were obtained from the following GenBank entries: C. elegans PP1, 1067062; D. melanogaster PP2A, 129338; P. vulgaris PP1, 1346765; H. sapiens PP2B/calcineurin, 180706; H. sapiens PP5, 2135921; R. rattus PP2A, 129333; S. pombe PP1, 130697.
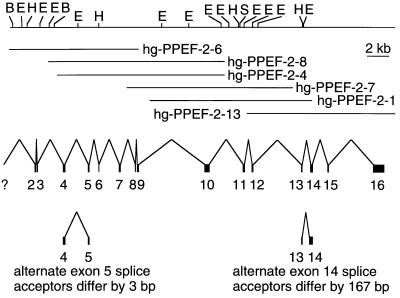
To determine the intron–exon structure of the human PPEF-2 gene, overlapping clones were isolated from a Sau3AI partial-digest human genomic DNA library. Fifteen exons, encoding amino acids 19–753, were identified within a genomic region of 33 kb (Fig. 2). The exon(s) encoding the first 18 aa has not been identified, and therefore, the 15 identified exons have been numbered 2–16. As seen in Fig. 1A, 8 of 11 intron locations in the Drosophila rdgC gene are precisely conserved in the human PPEF-2 gene, and two others are within several codons of their human counterparts. Introns 13 and 15 lie at corresponding locations in the center of imperfect and perfect matches to the EF-hand consensus, respectively, suggesting that this region evolved by exon duplication. The human PPEF-2 gene maps to chromosome 4 as determined by Southern blot hybridization (Oncor) and PCR amplification of DNA from panels of human–rodent hybrid cell lines (data not shown).
Figure 2.
Structures of the human PPEF-2 gene, mRNA, and splice variants. (Top) Restriction map encompassing 36 kb of human genomic DNA derived from the six lambda genomic clones shown below it. (Middle) The locations of the 15 identified exons are shown. The exon(s) encoding the first 18 aa has not been isolated; the exon encoding amino acids 19–61 has been designated exon 2. (Bottom) Two alternate splicing events are shown below the consensus mRNA structure. B, BamHI; E, EcoRI; H, HindIII; S, SalI.
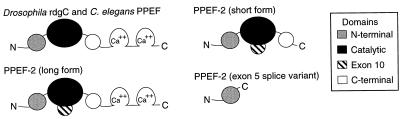
Two patterns of alternative splicing were identified by comparing the structures of multiple human PPEF-2 cDNA clones with the intron–exon structure of the PPEF-2 gene (Figs. 1, 2, 3). The 5′ end of exon 5 begins at either of two alternate acceptor sites that differ by three nucleotides. Use of the upstream acceptor results in the insertion of a stop codon (TAG) at amino acid 139, corresponding to the junction between an amino-terminal domain that is PPEF family-specific and the central catalytic core. Among five cDNA clones encompassing this region, two utilize the first exon 5 acceptor site and three utilize the second. Two alternate acceptor sites are present in exon 14, and these differ by 167 nt. The reading frame that results from use of the upstream splice acceptor site ends after 13 codons and generates a predicted protein of 598 aa that lacks the two carboxy-terminal EF-hand domains; we will refer to this short form of PPEF-2 as PPEF-2(S) (Figs. 1B and 3). The exon 14 reading frame that results from use of the downstream splice acceptor site continues uninterrupted through exons 14 and 15, and ends within exon 16 to generate a long form of PPEF-2, as depicted in Fig. 1A. This form, which we will refer to as PPEF-2(L), corresponds closely in overall structure to the other PPEF family members (see below). Among 24 cDNA clones that encompass exon 14, 7 encode PPEF-2(L) and 17 encode PPEF-2(S).
Figure 3.
Schematic diagram of PPEF protein structures. The central catalytic domain is deduced by a comparison between the PPEF subfamily and the primary sequences and three-dimensional structures of PP1 and PP2A/calcineurin. Sequences encoded by exon 10 of human PPEF-2 are presumed to protrude from the surface of the globular catalytic core.
Other PPEF Family Members.
Four PPEF-like sequences were identified in searches of the GenBank database (May 1997 release). Two expressed sequence tags (ESTs), obtained from human pineal and day 19.5 mouse embryo cDNA libraries (accession nos. AA069340 and AA059886), are perfect matches to the regions encoding the extreme carboxy termini of human and mouse PPEF-2, respectively. A third EST from a human fetal brain cDNA library (accession no. D44816) has a lower, but still significant degree of homology to the carboxy-terminal 80 aa of Drosophila rdgC and human and mouse PPEF-2. The sequences of two partial-length human retina cDNA clones isolated using the D44816 EF-hand region as a hybridization probe together with the sequence obtained from a 5′ RACE PCR product using human fetal brain cDNA as template reveal an ORF of 653 aa with 36% and 44% identity to Drosophila rdgC and human PPEF-2, respectively (Fig. 1A). While the present work was in progress, this sequence, referred to as PPEF, was independently identified and characterized by Montini et al. (31) as part of a comprehensive survey of genes encoded on the human X chromosome. We will refer to it hereafter as PPEF-1.
A sequence encoding the carboxy-terminal two-thirds of a PPEF-like protein has recently been identified by R. Waterston and colleagues in C. elegans genomic DNA (ORF F23H11.8). The missing 5′ region was obtained by 5′ RACE PCR and, together with the genomic sequence, reveals an ORF of 707 aa bearing all of the hallmarks of a PPEF family member (Figs. 1A and 3). This sequence, which will be referred to as C. elegans PPEF, has 34, 24, and 37% amino acid identity to Drosophila rdgC, human PPEF-2, and human PPEF-1, respectively. No other PPEF-like proteins are encoded in the 60% of the C. elegans genome determined to date.
Alignment of Drosophila rdgC, human and mouse PPEF-2, human PPEF-1, and C. elegans PPEF reveals a high degree of conservation in both spacing and amino acid identity throughout the 220 carboxy-terminal aa that lie outside of the core catalytic domain. This region in the PPEF subfamily shows little or no homology to the sequences of any other protein phosphatases. The alignment also reveals a region that is highly variable in both length and sequence inserted within the catalytic core beginning at amino acid 311 in human PPEF-2. This sequence is encoded by exon 10 in the human PPEF-2 gene and by exon 8 in the Drosophila rdgC gene (Fig. 3). It varies in size from 18 and 21 aa in Drosophila rdgC and C. elegans PPEF, respectively, to 47, 125, and 129 aa in human PPEF-1, human PPEF-2, and mouse PPEF-2, respectively. Comparison of the surrounding sequences with the sequences and three-dimensional structures of PP1 and PP2B/calcineurin suggest that this variable region would protrude from a surface loop that links α-helices 8 and 9 as defined in the PP2B/calcineurin structure (29, 30).
To assess the degree to which the PPEF branch has diverged from other branches of the PPP family of protein serine/threonine phosphatases, a dendrogram was calculated based on amino acid identity (Fig. 1D). The dendrogram shows that members of the PPEF subfamily cluster in a distinct branch.
Expression of PPEF-2 in Photoreceptors and Pinealocytes.
The tissue distribution of PPEF-2 transcripts was examined by Northern blot hybridization of total RNA from a variety of adult rat tissues. A major PPEF-2 transcript of approximately 3.7 kb is readily detected in retina RNA, whereas RNAs from other tissues show no detectable hybridization (Fig. 4). The transcript size is consistent with the predicted sizes of 3.3 and 3.4 kb (not including the poly(A) tail) of the mouse and human PPEF-2(L) mRNAs, respectively, that were deduced from sequences of cDNA clones that include apparently complete 3′ noncoding regions.
Figure 4.
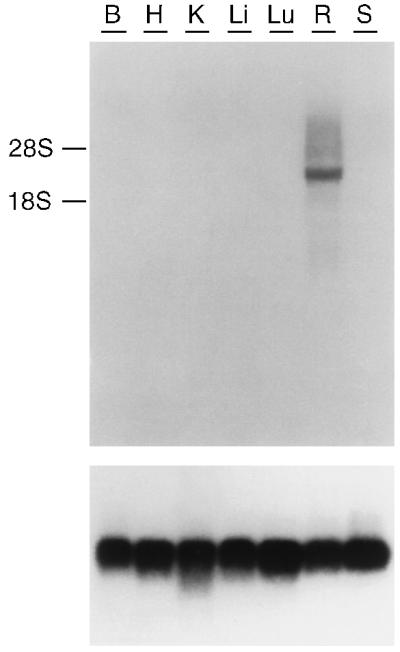
Size and distribution of PPEF-2 transcripts in adult rat tissues. A blot of total RNA from the indicated tissues was hybridized with a 1-kb human PPEF-2 cDNA probe encompassing exons 1–9 (Upper). Migration of the 28S and 18S ribosomal RNAs is shown. A major retina-specific band of approximately 3.7 kb is evident. As a control, the same blot was probed with a rat cDNA encoding ribosomal protein S26 (Lower; ref. 49). B, brain; H, heart; K, kidney; Li, liver; Lu, lung; R, retina; S, spleen.
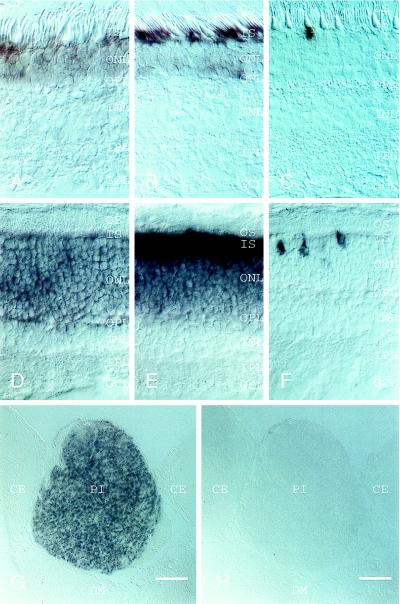
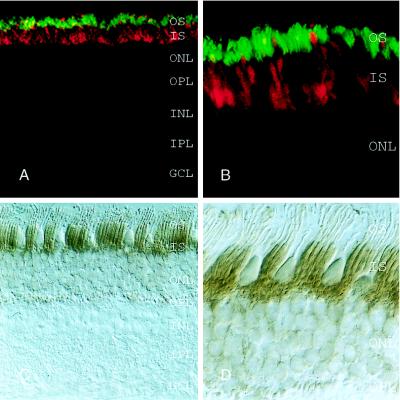
To further define the cell types in which PPEF-2 is expressed, rat, mouse, and macaque retinas were examined by in situ hybridization (Fig. 6; data not shown). In retinas from all three species, PPEF-2 transcripts are present in photoreceptors. With the exception of weak hybridization to occasional cells within the inner nuclear layer in rodent retinas, no other retinal cells contain PPEF-2 transcripts. Although the uniform PPEF-2 hybridization pattern in the outer nuclear layer clearly indicates expression in rods, the relative paucity of cones in these species and the partial superposition of rod and cone hybridization signals in 20-μm sections does not allow us to draw a clear conclusion from these data regarding PPEF-2 expression in cones. Interestingly, PPEF-2 transcripts do not cluster in the inner segment region of rods as do rhodopsin transcripts (compare Fig. 6 A with B, and D with E).
Figure 6.
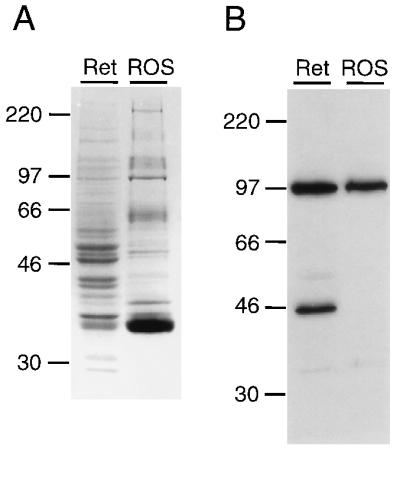
Identification of PPEF-2(L) by immunoblotting of proteins from bovine retina (Ret) or rod outer segments (ROS). Protein (25 μg) was loaded in each lane and visualized with Coomassie Brilliant Blue R-250 (A), or immunoblotted and probed with affinity-purified rabbit antibodies directed against the carboxy-terminal 212 residues of PPEF-2(L) (B). Molecular masses of protein size standards are shown.
The mammalian pineal expresses a subset of genes whose products function in retinal photoreception, including interphotoreceptor retinoid binding protein (32), rhodopsin kinase (33), and arrestin (34), and during development pinealocytes display a photoreceptor-like morphology (35). As determined by in situ hybridization, the PPEF-2 gene is also expressed in the adult rat pineal but not in other brain structures (Fig. 6G). By contrast, the rhodopsin gene is not detectably expressed in the rat pineal when analyzed under identical conditions (Fig. 6H). We presume that the pineal PPEF-2 transcript was not detected in total rat brain RNA by Northern blotting (Fig. 5) because the pineal occupies less than 1% of the volume of the brain.
Figure 5.
In situ hybridization localizes PPEF-2 transcripts to retinal photoreceptors and pinealocytes. (A–C) Macaque retina. (D–F) Rat retina. (G and H) Coronal section of rat brain through the pineal and surrounding structures. Sections were hybridized with the following probes: human PPEF-2 (A); mouse PPEF-2 (D and G); macaque rhodopsin (B, E, and H); and mouse blue cone pigment (C and F). In the retina, PPEF-2 transcripts are confined to photoreceptor cells. PPEF-2 transcripts are also readily detected in the pineal, whereas rhodopsin transcripts are detectable only in the retina. The rat retina and pineal sections were present on the same slide so that hybridization to these two tissues was performed under identical conditions. OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer; PI, pineal; CE, cerebrum; DM, dorsal midbrain.
Localization of PPEF-2(L) to Rod Inner Segments.
The specific expression of PPEF-2 in retinal photoreceptors and the pineal suggests that PPEF-2 may play a role in vertebrate phototransduction, as indicated for Drosophila rdgC (5, 16). As an initial test of this possibility we determined whether PPEF-2 was enriched in rod outer segments, the site of phototransduction. Rabbit polyclonal antibodies directed against a fusion protein containing the carboxy-terminal 208 aa of human PPEF-2(L) were affinity-purified and used to visualize this form of PPEF-2 on immunoblots containing equal amounts of protein either from bovine rod outer segments isolated by sucrose gradient centrifugation or from crude extracts of bovine retina (Fig. 6). These antibodies do not recognize the 43-aa region that is shared between PPEF-2(S) and the PPEF-2(L) fusion protein used for immunization (data not shown). The immunoblot shows a band in samples from both rod outer segments and total retina with an apparent mass of 95 kDa, which is consistent with the predicted molecular mass of 86.4 kDa for human PPEF-2(L). A second band with an apparent mass of 45 kDa is consistently seen only in the retina extract and may represent a cross-reacting protein or a breakdown product of the larger band. This experiment shows that PPEF-2(L) is at least 2-fold more abundant per milligram of protein in the total retina extract than it is in rod outer segments. By contrast, rhodopsin, a rod outer segment-specific protein, is at least 10-fold more abundant per milligram of protein in the rod outer segment preparation than it is in the total retina extract (36). Given that PPEF-2 transcripts and, therefore, PPEF-2 protein is localized to photoreceptors and that approximately two-thirds of the protein extract from whole retina is derived from retinal cell types other than photoreceptors, it is likely that the vast majority of PPEF-2(L) is not present in photoreceptor outer segments. The small fraction present in the rod outer segment preparation may reflect low levels of PPEF-2(L) in the rod outer segments or may derive from contamination by attached inner segments or other photoreceptor cell fragments.
Attempts to visualize PPEF-2(S) in retina using affinity-purified rabbit antibodies directed against a fusion protein containing the carboxy-terminal 57 aa of PPEF-2(S) have thus far failed despite the comparable abundance in the human retina of the mRNA species encoding PPEF-2(L) and PPEF-2(S). The existence of PPEF-2(S) in vivo remains to be determined.
Immunohistochemistry in bovine, cat, and macaque retinas using the antibodies described above localizes PPEF-2(L) to rod inner segments and, to a lesser extent, to the presynaptic terminals (Fig. 7; data not shown), consistent with the interpretation of the immunoblotting data outlined above. In the macaque retina, cones are readily distinguished from rods by their larger inner segments and smaller outer segments. As seen in Fig. 7 C and D, rod inner segments are clearly stained with anti-PPEF-2(L) antibody, whereas cone inner segments show little or no immunostaining.
Figure 7.
PPEF-2(L) is localized to rod inner segments. Immunolabeling was performed with affinity-purified rabbit antibodies directed against the carboxy-terminal 212 residues of PPEF-2(L). (A and B) One-micrometer optical sections showing immunofluorescent double-labeling of bovine retina with anti-PPEF-2 antibodies (Texas Red) and mouse mAb PMc1D1 against the rod outer segment cGMP-gated channel (fluorescein). (C and D) Immunoperoxidase staining of macaque retina with anti-PPEF-2 antibodies. PPEF-2(L) is confined to the inner segments in both bovine and macaque retinas. In the macaque retina, immunostaining is confined to the narrow rod inner segments; the large cone inner segments are not stained. (A and C) Low magnification. (B and D) High magnification.
DISCUSSION
The experiments reported here show that homologues of the Drosophila rdgC phosphatase exist both in mammals and in the simple multicellular worm C. elegans. Thus, the PPEF subfamily of protein serine/threonine phosphatases represents a conserved branch of the PPP family that is likely to be present throughout the animal kingdom. One distinctive structural feature of PPEF family members is their large size (653–757 aa) as compared with other PPP phosphatases (approximately 320, 310, 525, and 500 aa for PP1, PP2A, PP2B/calcineurin, and PP5, respectively). A second feature is the presence of at least two EF-hand motifs near the carboxy terminus, strongly suggesting that the enzymatic activity of the PPEF family is regulated by intracellular calcium. A protein serine/threonine phosphatase from Arabidopsis that is involved in absisic acid signal transduction has amino-terminal EF-hands, but it is a member of the PP2C family of phosphatases (37, 38).
Calcium regulation of protein serine/threonine phosphatase activity has been extensively studied in PP2B/calcineurin, which is regulated both by calmodulin and by calcineurin B, a calmodulin-like protein (39). Calmodulin and calcineurin B bind to distinct sites in the carboxy-terminal regulatory domain of the catalytic subunit, thereby displacing an autoinhibitory domain from the enzyme active site (40–42). This precedent suggests that the carboxy-terminal 200 aa of the PPEF phosphatases may also comprise a regulatory domain that is modulated by calcium binding to the carboxy-terminal EF-hand domain and/or by anchoring proteins, inhibitors, or activators (43, 44).
The specific localization of PPEF-2 transcripts in retinal rods and in the pineal is in contrast to the broader tissue distribution typically seen for other PPP phosphatases (44) and strongly suggests that, like Drosophila rdgC, PPEF-2 plays a specialized role in the visual system. However, the high concentration of PPEF-2(L) in the photoreceptor inner segment relative to the outer segment is inconsistent with the hypothesis that this protein acts in the phototransduction cascade and, in particular, that it dephosphorylates photoactivated rhodopsin (or opsin in the mammalian system), as has been proposed for rdgC in Drosophila photoreceptors (5, 16). Current evidence suggests that a PP2A catalytic subunit effects dephosphorylation of vertebrate phospho-opsin (45, 46) and other G protein-coupled receptors (47), although one recent study indicates that bovine rod outer segments contain both calcium-independent and calcium-dependent dephosphorylation activities (48). If mammalian PPEF-2 and Drosophila rdgC are assumed to act on homologous substrates, then the data could be reconciled by postulating that PPEF-2(S)—if it exists in vivo—or a small amount of PPEF-2(L) is present in rod outer segments and is sufficient to dephosphorylate rhodopsin. Alternately, PPEF phosphatases may act on substrates other than rhodopsin, and in Drosophila this could indirectly promote rhodopsin dephosphorylation. We note that if Drosophila rdgC and mammalian PPEF-2 exhibit similar modes of regulation by calcium, then photoactivation will have opposite effects on phosphatase activity in these two systems because cytosolic calcium rises in Drosophila photoreceptors and falls in mammalian photoreceptors following light exposure. The localization of PPEF-2(L) to the cell body of mammalian photoreceptors suggests that this protein may function as a calcium-sensing regulator of ionic currents, energy production, or synaptic transmission.
The recent demonstration that PPEF-1 transcripts in the developing mouse are restricted to primary somatosensory neurons and to the inner ear (31) suggests that PPEF-2 and PPEF-1 may play analogous roles in different sensory systems. We speculate that in auditory, visual, and somatosensory neurons, modulation of sensory inputs may proceed via analogous cycles of protein serine/threonine phosphorylation and dephosphorylation, with the latter mediated by PPEF family members in a calcium-dependent manner. The existence of a PPEF sequence in C. elegans is consistent with this proposed conservation of function and suggests that a genetic analysis of the C. elegans gene may shed light on the function of the PPEF family in higher organisms.
Acknowledgments
We thank R. Molday for mAbs 1D4 and PMc1D1; A. Rattner for the gift of the rat RNA blot; Y. Wang for the human genomic library; C. Riley, C. Davenport, J. Ptak, and M. Kazienko for oligonucleotide synthesis; M. Delannoy for assistance with confocal microscopy; H. Zhou and S. Hendry for macaque retinae; and D. Valle and Y. Wang for comments on the manuscript. This work was supported by the Ruth and Milton Steinbach Fund and the Howard Hughes Medical Institute.
ABBREVIATIONS
- PPEF-1 and PPEF-2
Protein Phosphatase with EF-hands-1 and 2
- rdgC
Drosophila retinal degeneration C
Footnotes
References
- 1.Stryer L. Annu Rev Neurosci. 1986;9:87–119. doi: 10.1146/annurev.ne.09.030186.000511. [DOI] [PubMed] [Google Scholar]
- 2.Yau K-W. Investigative Ophthalmol Vis Sci. 1994;35:9–32. [PubMed] [Google Scholar]
- 3.Ranganathan R, Malicka D M, Zuker C S. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 4.Wilden U, Hall S W, Kuhn H. Proc Natl Acad Sci USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byk T, Bar-Yaacov M, Doza Y N, Minke B, Selinger Z. Proc Natl Acad Sci USA. 1993;90:1907–1911. doi: 10.1073/pnas.90.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pak W L. Inv Ophthalmol Vis Sci. 1995;36:2340–2357. [PubMed] [Google Scholar]
- 7.Dryja T P, Li T. Human Mol Genet. 1995;4:1739–1743. doi: 10.1093/hmg/4.suppl_1.1739. [DOI] [PubMed] [Google Scholar]
- 8.Sung C-H, Schneider B G, Agarwal N, Papermaster D S, Nathans J. Proc Natl Acad Sci USA. 1991;88:8840–D8844. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaushal S, Khorana H G. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 10.Kurada P, O’Tousa J E. Neuron. 1995;14:571–579. doi: 10.1016/0896-6273(95)90313-5. [DOI] [PubMed] [Google Scholar]
- 11.Colley N J, Cassill J A, Baker E K, Zuker C S. Proc Natl Acad Sci USA. 1995;92:3070–3074. doi: 10.1073/pnas.92.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolph P J, Ranganathan R, Colley N J, Hardy R W, Socolich M, Zuker C S. Science. 1993;260:1910–1916. doi: 10.1126/science.8316831. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. Nat Genet. 1995;10:360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- 14.Steele F, O’Tousa J E. Neuron. 1990;4:883–890. doi: 10.1016/0896-6273(90)90141-2. [DOI] [PubMed] [Google Scholar]
- 15.Steele F R, Washburn T, Rieger R, O’Tousa J E. Cell. 1992;69:669–676. doi: 10.1016/0092-8674(92)90230-a. [DOI] [PubMed] [Google Scholar]
- 16.Vinós J, Jalink K, Hardy R W, Britt S G, Zuker C S. Science. 1997;277:687–690. doi: 10.1126/science.277.5326.687. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Macke J P, Abella B S, Andreasson K, Worley P, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. J Biol Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 18.Nathans J, Thomas D, Hogness D S. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 19.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blumenthal T, Steward K. In: C. Elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors; Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Coldspring Harbor, NY: Cold Spring Harbor Lab. Press; 1997. pp. 117–146. [Google Scholar]
- 21.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 23.Chiu M I, Zack D J, Wang Y, Nathans J. Genomics. 1994;21:440–443. doi: 10.1006/geno.1994.1292. [DOI] [PubMed] [Google Scholar]
- 24.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 25.Papermaster D S. Methods Enzymol. 1982;81:48–52. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 26.Cook N J, Molday L L, Reid D, Kaupp U B, Molday R S. J Biol Chem. 1989;264:6996–6999. [PubMed] [Google Scholar]
- 27.Kretsinger R H. Cold Spring Harbor Symp Quant Biol. 1987;52:499–510. doi: 10.1101/sqb.1987.052.01.057. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg J, Huang H B, Kwon Y G, Greengard P, Nairn A C, Kuriyan J. Nature (London) 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 29.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 30.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, Gastinel L N, Habuka N, Chen X, Maldonado F, Barker J E, Bacquet R, Villafranca J E. Nature (London) 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 31.Montini E, Rugarli E I, Van de Vosse E, Andolfi G, Mariani M, Puca A A, Consalez G G, den Dunnen J T, Ballabio A, Franco B. Human Mol Genet. 1997;6:1137–1145. doi: 10.1093/hmg/6.7.1137. [DOI] [PubMed] [Google Scholar]
- 32.Bridges C D B, Foster R G, Landers R A, Fong S L. Vis Res. 1987;27:2049–2060. doi: 10.1016/0042-6989(87)90119-2. [DOI] [PubMed] [Google Scholar]
- 33.Palczewski K, Carruth M E, Adamus G, McDowell J H, Hargrave P. Vis Res. 1990;30:1129–1137. doi: 10.1016/0042-6989(90)90170-p. [DOI] [PubMed] [Google Scholar]
- 34.Abe T, Shinohara T. Exp Eye Res. 1990;51:111–112. doi: 10.1016/0014-4835(90)90178-w. [DOI] [PubMed] [Google Scholar]
- 35.Zimmerman B L, Tso M O M. J Cell Biol. 1975;66:60–75. doi: 10.1083/jcb.66.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun H, Nathans J. Nat Genet. 1997;17:15–16. doi: 10.1038/ng0997-15. [DOI] [PubMed] [Google Scholar]
- 37.Leung J, Bouvier-Durand M, Morris P-C, Guerrier D, Chefdor F, Giraudat J. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 38.Meyer K, Leube M P, Grill E. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 39.Stemmer P, Klee C B. Curr Opinion Neurobiol. 1991;1:53–64. doi: 10.1016/0959-4388(91)90010-5. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto Y, Perrino B A, Soderling T R. J Biol Chem. 1990;265:1924–1927. [PubMed] [Google Scholar]
- 41.Parsons J N, Wiederrecht G J, Salowe S, Burbaum J J, Rokosz L L, Kincaid R L, O’Keefe S J. J Biol Chem. 1994;269:19610–19616. [PubMed] [Google Scholar]
- 42.Perrino B A, Ng L Y, Soderling T R. J Biol Chem. 1995;270:340–346. doi: 10.1074/jbc.270.1.340. [DOI] [PubMed] [Google Scholar]
- 43.Hubbard M J, Cohen P. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 44.Shenolikar S. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 45.Fowles C, Akhtar M, Cohen P. Biochemistry. 1989;28:9385–9391. doi: 10.1021/bi00450a020. [DOI] [PubMed] [Google Scholar]
- 46.Palczewski K, Hargrave P A, McDowell J H, Ingebritsen T S. Biochemistry. 1989;28:415–419. doi: 10.1021/bi00428a001. [DOI] [PubMed] [Google Scholar]
- 47.Pitcher J A, Payne E S, Csortos C, DePaoli-Roach A A, Lefkowitz R J. Proc Natl Acad Sci USA. 1995;92:8343–8347. doi: 10.1073/pnas.92.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutuzov M A, Bennett N. Eur J Biochem. 1996;238:613–622. doi: 10.1111/j.1432-1033.1996.0613w.x. [DOI] [PubMed] [Google Scholar]
- 49.Kuwano Y, Nakanishi O, Nabeshima Y-I, Tanaka T, Ogata K. J Biochem (Tokyo) 1985;97:983–992. doi: 10.1093/oxfordjournals.jbchem.a135175. [DOI] [PubMed] [Google Scholar]