Abstract
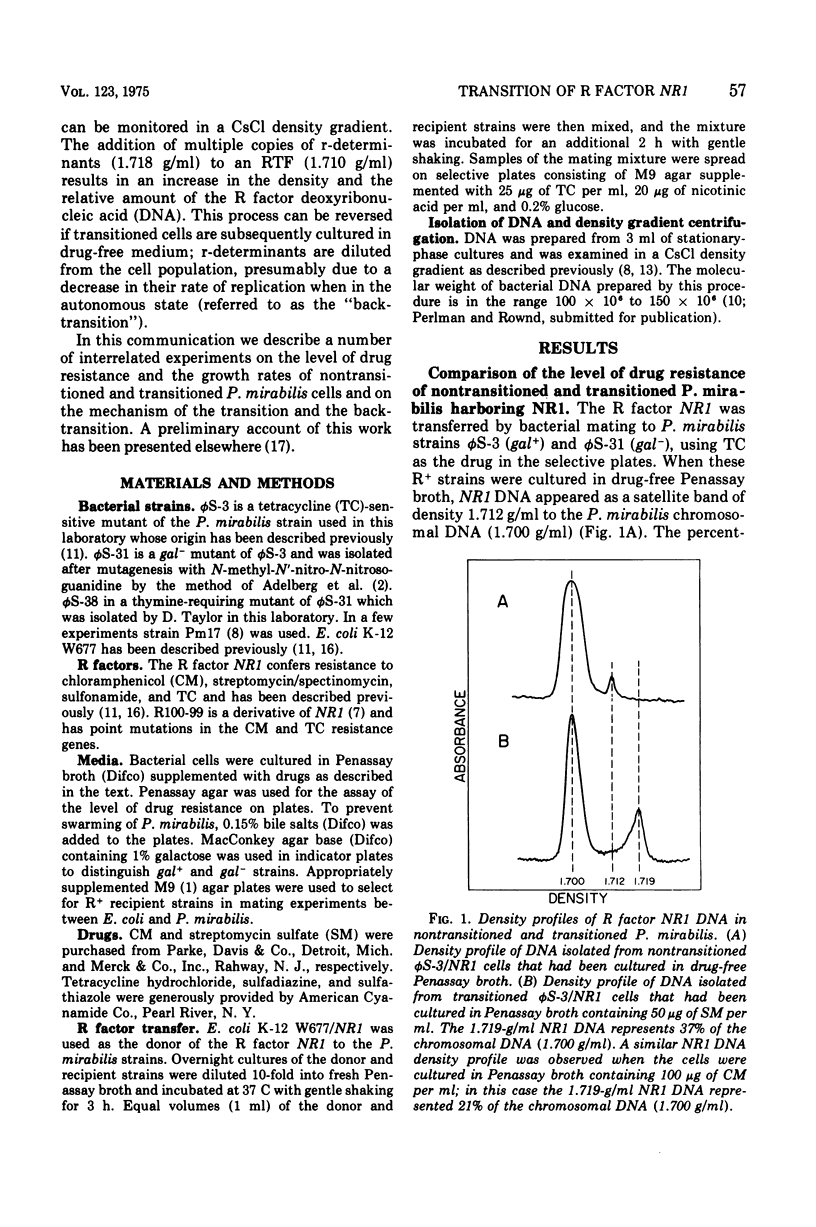
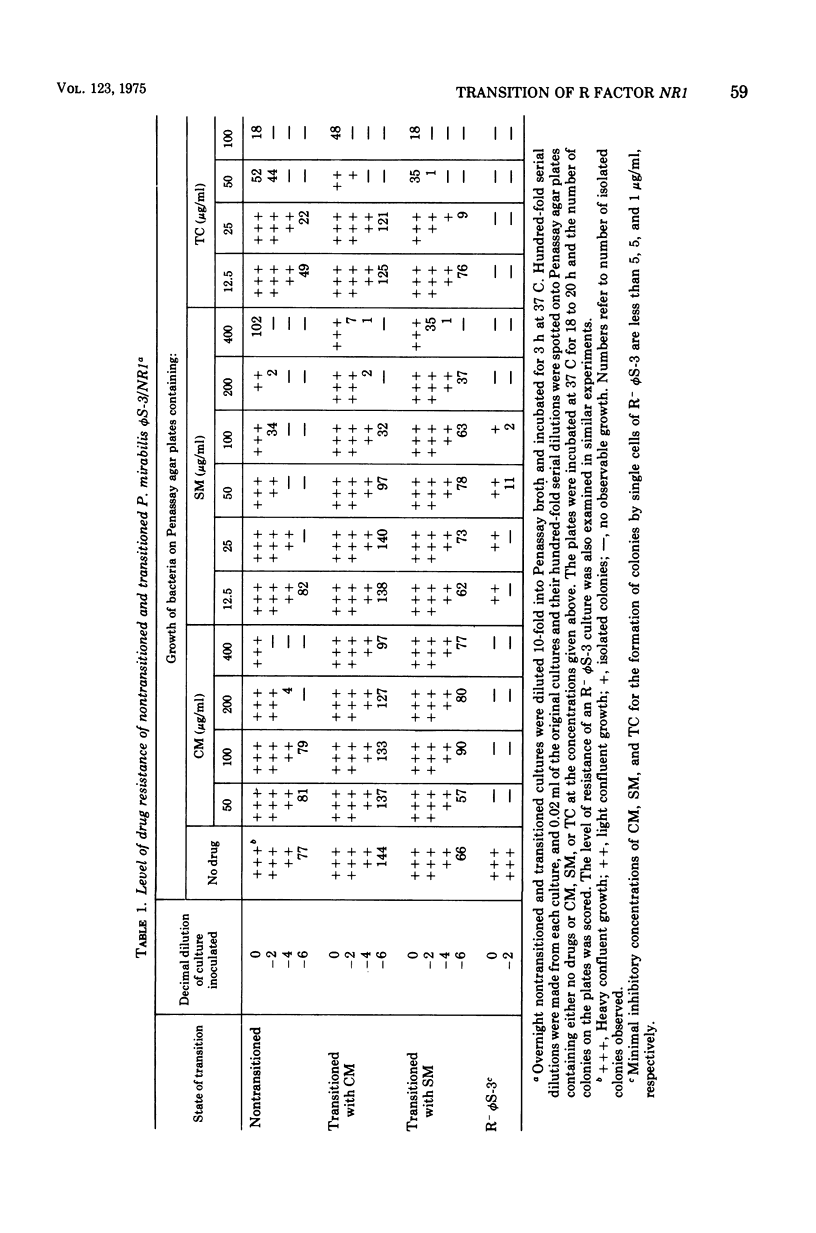
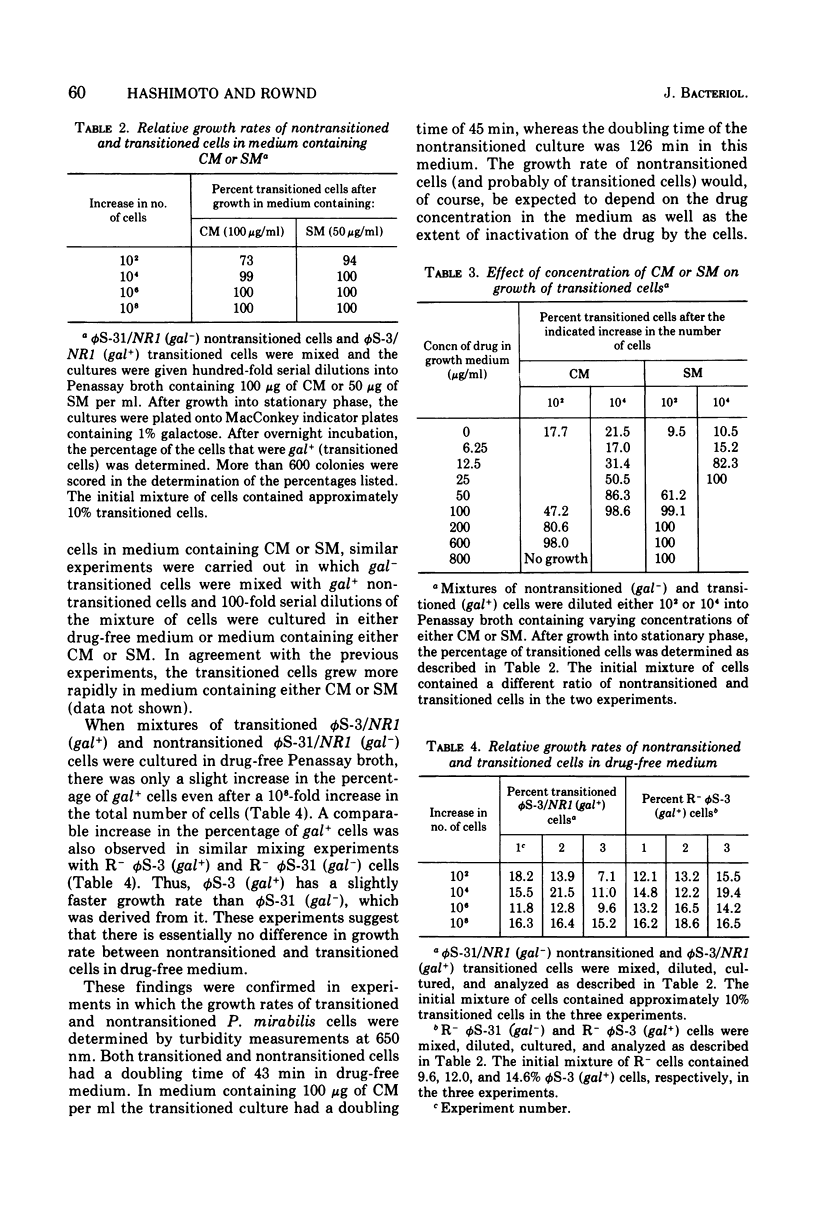
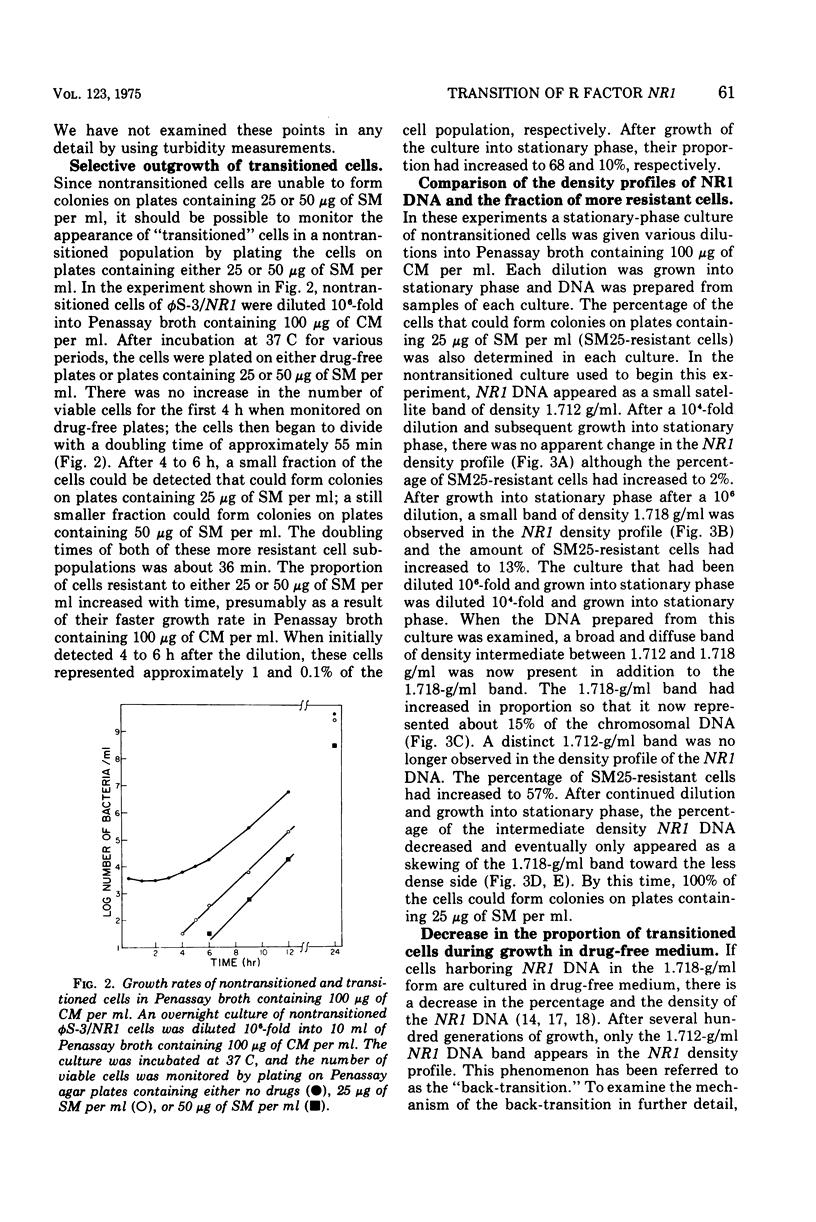
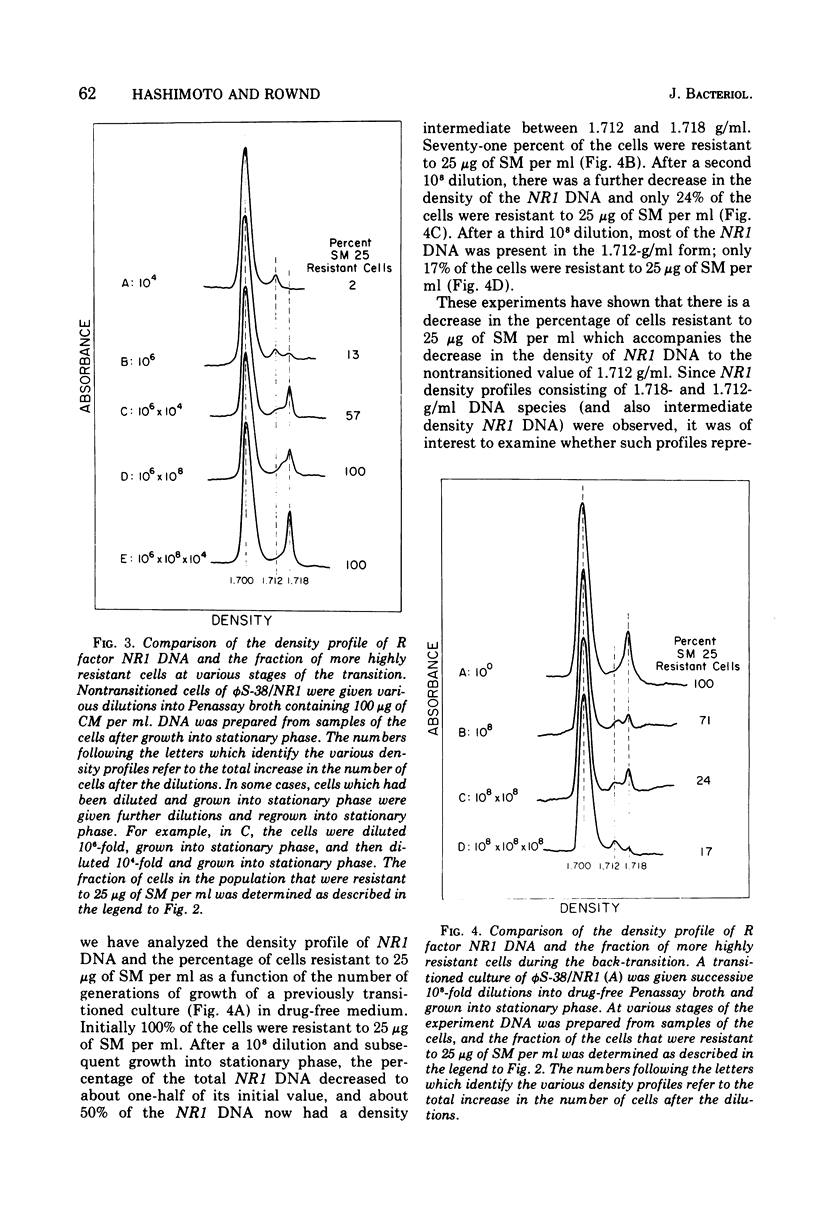
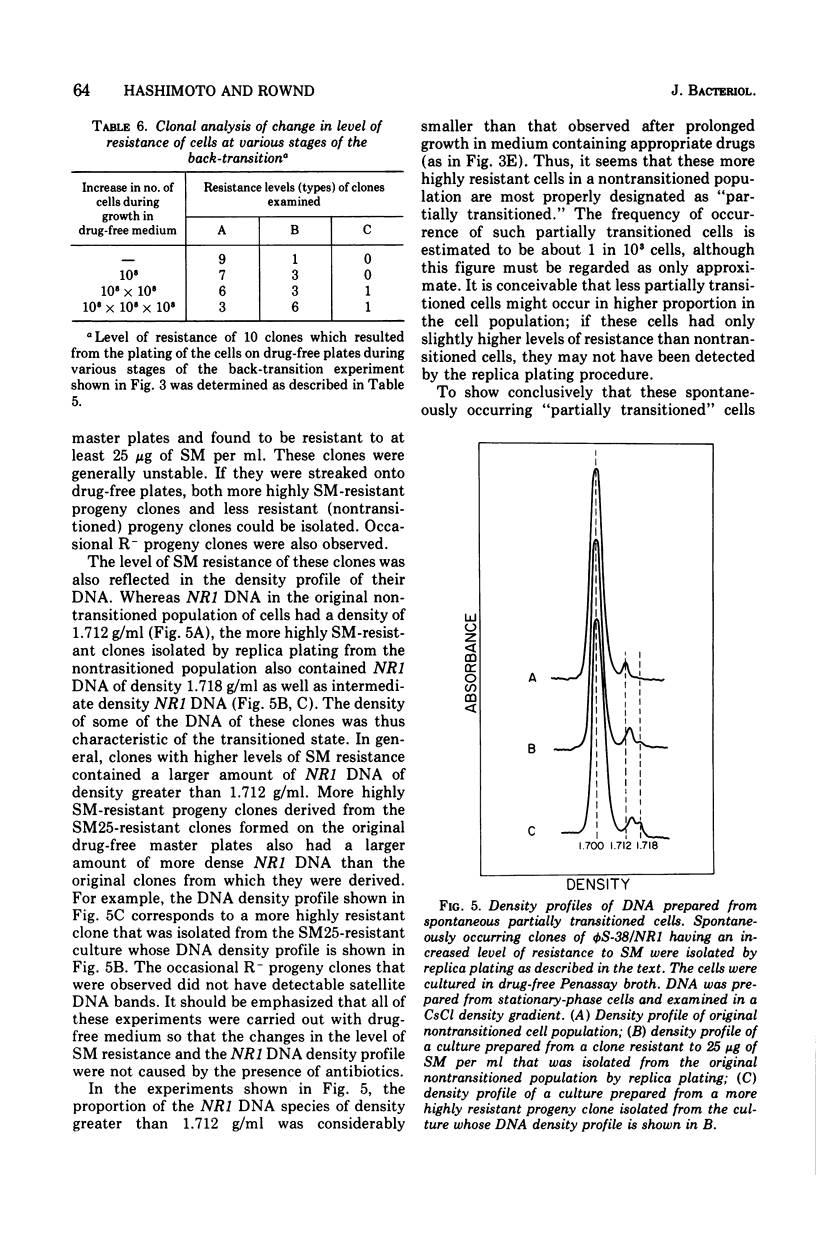
When Proteus mirabilis harboring the R factor NR1 is cultured in Penassay broth containing 100 mug of chloramphenicol (CM) per ml, there is an amplification in the number of copies of the r-determinants per cell. Under these conditions, R factors harboring multiple tandem sequences of r-determinants are formed. Autonomous poly-f-determinants consisting of multiple copies of r-determinants are also formed. This phenomenon has been referred to as the "transition". Transitioned cells have considerably higher levels of resistance to CM and streptomycin (SM), but not to tetracycline (TC), than do nontransitioned cells and grow more rapidly in medium containing either CM or SM. There is essentially no difference in growth rates between transitioned and nontransitioned cells in drug-free medium. The higher level of resistance of transitioned cells to SM has made it possible to investigate the mechanism of the transition. Using replica plating, it has been possible to isolate spontaneously occurring transitioned cells from a nontransitioned population which appear to outgrow the nontransitioned cells during growth in medium containing 100 mug of CM per ml. If transiitoned cells are subsequently cultured in drug-free medium, the cells return gradually to the nontransitioned state, which has been referred to as the "back-transition was monitored by examining the level of resisitance of the cells to SM. In both situations the cell populations were found to be heterogeneous, consisting of a mixture of nontransitioned and transitioned cells. Under the conditions of our experiments, the transition appeared to be due to the more rapid growth of a minor fraction of spontaneously occurring transitioned cells which outgrew the remainder of cells in the population. To obtain the transition, the drug resistance gene must reside on the r-determinants component of the R factor. The transition did not take place when the cells were cultured in medium containing high concentrations of TC. This indicates that the TC resistance genes reside on the resistance transfer factor component of the R factor, which is in agreement with physical studies on R factor deoxyribonucleic acid.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S. The ecology of transferable drug resistance in the enterobacteria. Annu Rev Microbiol. 1968;22:131–180. doi: 10.1146/annurev.mi.22.100168.001023. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Miller C. A. Non-chromosomal antibiotic resistance in bacteria. II. Molecular nature of R-factors isolated from Proteus mirabilis and Escherichia coli. J Mol Biol. 1970 Jun 28;50(3):671–687. doi: 10.1016/0022-2836(70)90092-6. [DOI] [PubMed] [Google Scholar]
- Franklin T. J., Rownd R. R-factor-mediated resistance to tetracycline in Proteus mirabilis. J Bacteriol. 1973 Jul;115(1):235–242. doi: 10.1128/jb.115.1.235-242.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Hirota Y. Gene recombination and segregation of resistance factor R in Escherichia coli. J Bacteriol. 1966 Jan;91(1):51–62. doi: 10.1128/jb.91.1.51-62.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu H., Rownd R. Replication of R-factors in Proteus mirabilis: replication under relaxed control. J Mol Biol. 1970 Aug;51(3):473–489. doi: 10.1016/0022-2836(70)90002-1. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Rownd R. Transition of the R factor R12 in Proteus mirabilis. J Bacteriol. 1974 Jun;118(3):867–879. doi: 10.1128/jb.118.3.867-879.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya R., Rownd R. Transduction of R factors by a Proteus mirabilis bacteriophage. J Bacteriol. 1971 Jun;106(3):773–783. doi: 10.1128/jb.106.3.773-783.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Kasamatsu H., Mickel S. The molecular nature and replication of drug resistance factors of the Enterobacteriaceae. Ann N Y Acad Sci. 1971 Jun 11;182:188–206. doi: 10.1111/j.1749-6632.1971.tb30656.x. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- Rownd R., Perlman D., Hashimoto H., Mickel S., Applebaum E., Taylor D. Dissociation and reassociation of the transfer factor and resistance determinants of R factors as a mechanism of gene amplification in bacteria. Johns Hopkins Med J Suppl. 1973;2:115–128. [PubMed] [Google Scholar]
- Rownd R. Replication of a bacterial episome under relaxed control. J Mol Biol. 1969 Sep 28;44(3):387–402. doi: 10.1016/0022-2836(69)90368-4. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. The problems of drug-resistant pathogenic bacteria. Comparative enzymology of chloramphenicol resistance. Ann N Y Acad Sci. 1971 Jun 11;182:234–242. doi: 10.1111/j.1749-6632.1971.tb30660.x. [DOI] [PubMed] [Google Scholar]
- WATANABE T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963 Mar;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]



