Abstract
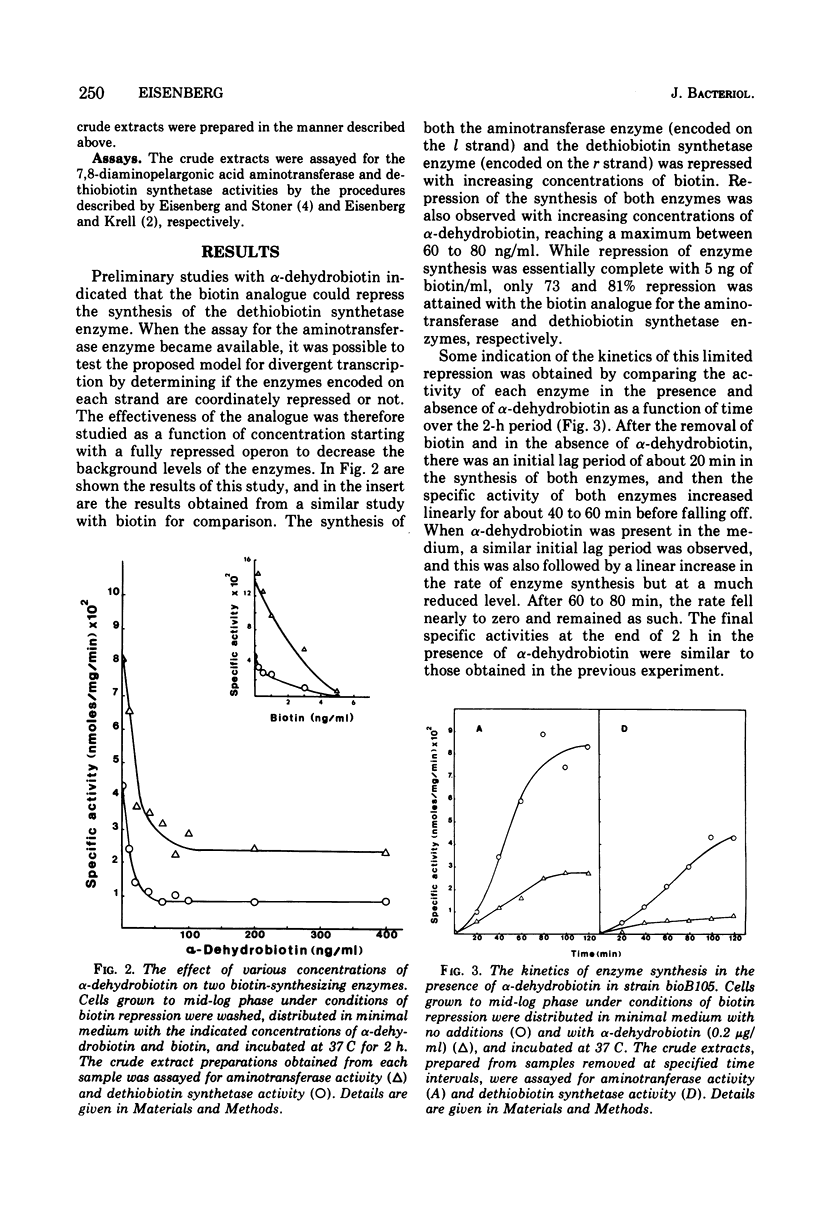
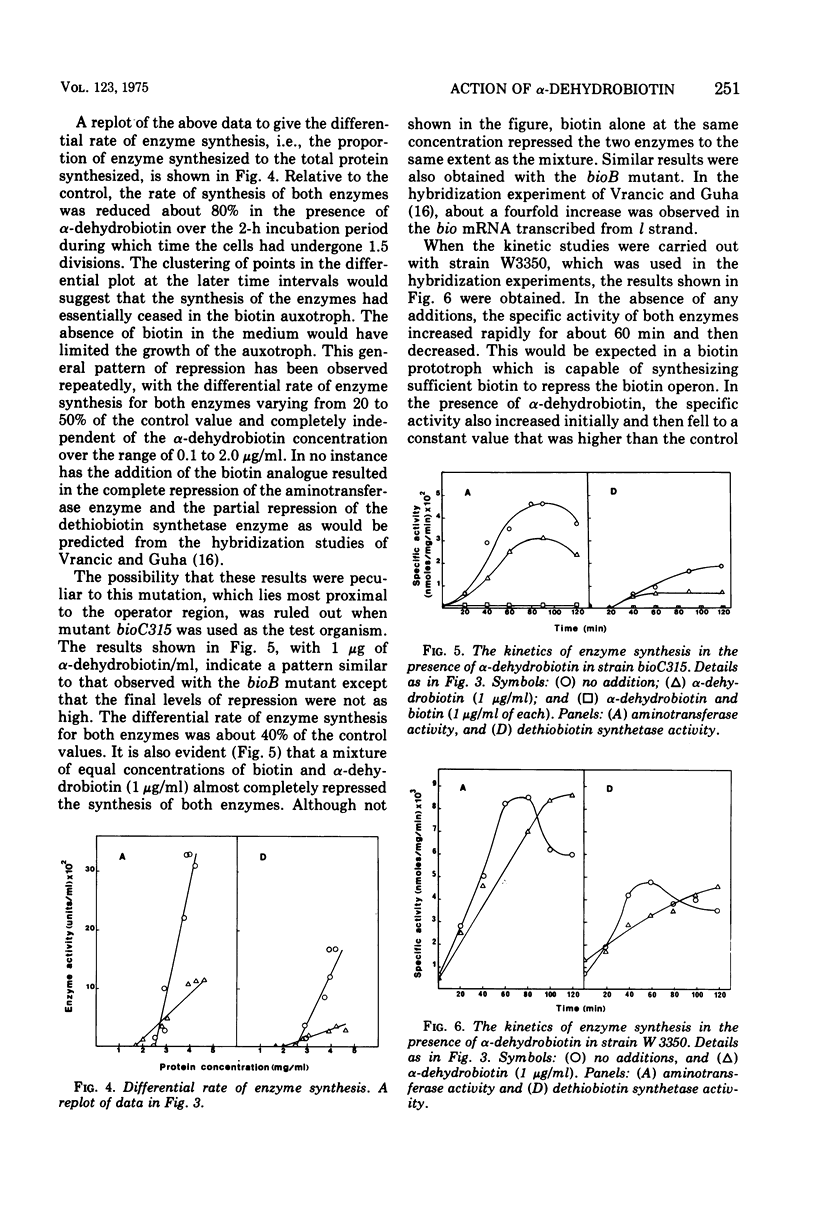
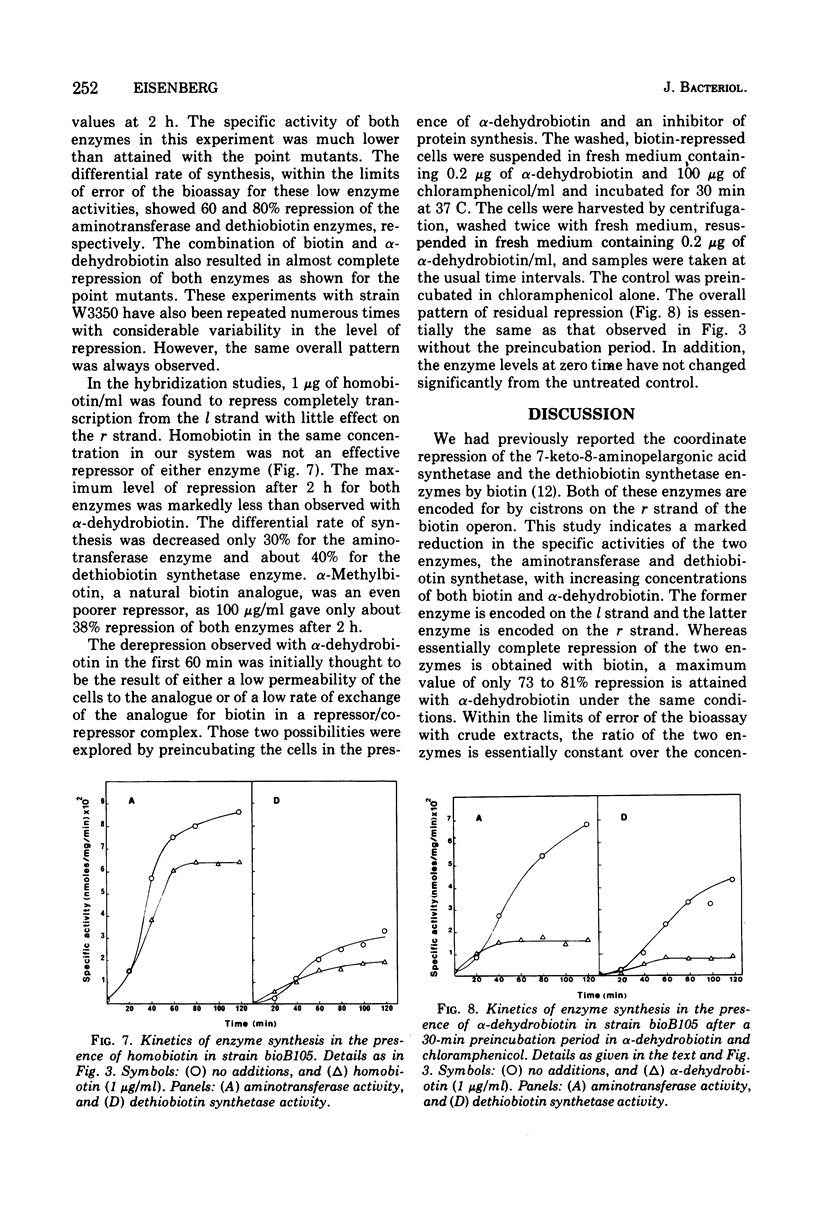
Alpha-Dehydrobiotin, like biotin, represses coordinately the 7,8-diaminopelargonic acid aminotransferase and the dethiobiotin synthetase enzymes that are encoded on the l and r strands, respectively, of the bioA operon. The rate of synthesis for both enzymes is inhibited about 80% in the presence of alpha-dehydrobiotin. Homobiotin and alpha-methylbiotin are less effective than alpha-dehydrobiotin in repressing the synthesis of the two enzymes. The selective repression of transcription from l and by alpha-dehydrobiotin and homobiotin, previously reported in hybridization experiments, is not observed at the enzyme level. A combination of equal concentrations of biotin and alpha-dehydrobiotin which was reported to enhance selectively the level of messenger ribonucleic acid transcribed from the l strand does not increase the rate of synthesis of the aminotransferase enzyme. Instead, the enzymes encoded on both strands are essentially completely inhibited as with biotin alone. Strain differences have been ruled out to account for the different results obtained by the two methodologies. Our evidence would suggest that alpha-dehydrobiotin acts like biotin, presumably as the co-repressor, in the repression of the bioA operon. The low rates of enzyme synthesis observed in the presence of the biotin analogue is the result of incomplete repression due to a lower affinity of either the analogue for the repressor or of the co-repressor/repressor complex for the operator. While our evidence would support the concept of a two promoter/operator complex, both would have to respond equally to biotin and its analogues. The evidence, however, does not rule out other possible alternative models for the regulation of the biotin operon.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Eisenberg M. A. Biotin: biogenesis, transport, and their regulation. Adv Enzymol Relat Areas Mol Biol. 1973;38:317–372. doi: 10.1002/9780470122839.ch7. [DOI] [PubMed] [Google Scholar]
- Eisenberg M. A., Krell K. Dethiobiotin synthesis from 7,8-diaminolargonic acid in cell-free extracts of a biotin auxotroph of Escherichia coli K-12. J Biol Chem. 1969 Oct 25;244(20):5503–5509. [PubMed] [Google Scholar]
- Eisenberg M. A., Krell K. Synthesis of desthiobiotin from 7,8-diaminopelargonic acid in biotin auxotrophs of Escherichia coli K-12. J Bacteriol. 1969 Jun;98(3):1227–1231. doi: 10.1128/jb.98.3.1227-1231.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg M. A., Stoner G. L. Biosynthesis of 7,8-diaminopelargonic acid, a biotin intermediate, from 7-keto-8-aminopelargonic acid and S-adenosyl-L-methionine. J Bacteriol. 1971 Dec;108(3):1135–1140. doi: 10.1128/jb.108.3.1135-1140.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenburg M. A., Mee B., Prakash O., Eisenburg M. R. Properties of alpha-dehydrobiotin-resistant mutants of Escherichia coli K-12. J Bacteriol. 1975 Apr;122(1):66–72. doi: 10.1128/jb.122.1.66-72.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A. Divergent orientation of transcription from the biotin locus of Escherichia coli. J Mol Biol. 1971 Feb 28;56(1):53–62. doi: 10.1016/0022-2836(71)90083-0. [DOI] [PubMed] [Google Scholar]
- Hanka L. J., Bergy M. E., Kelly R. B. Naturally occurring antimetabolite antibiotic related to biotin. Science. 1966 Dec 30;154(3757):1667–1668. doi: 10.1126/science.154.3757.1667. [DOI] [PubMed] [Google Scholar]
- Hanka L. J., Martin D. G., Reineke L. M. Two new antimetabolites of biotin: alpha-methyldethiobiotin and alpha-methylbiotin. Antimicrob Agents Chemother. 1972 Feb;1(2):135–138. doi: 10.1128/aac.1.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanka L. J., Reineke L. M., Martin D. G. Biological studies with alpha-dehydrobiotin. J Bacteriol. 1969 Oct;100(1):42–46. doi: 10.1128/jb.100.1.42-46.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketner G., Campbell A. A deletion mutation placing the galactokinase gene of Escherichia coli under control of the biotin promoter. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2698–2702. doi: 10.1073/pnas.71.7.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell K., Gottesman M. E., Parks J. S., Eisenberg M. A. Escape synthesis of the biotin operon in induced lambda b-2 lysogens. J Mol Biol. 1972 Jul 14;68(1):69–82. doi: 10.1016/0022-2836(72)90263-x. [DOI] [PubMed] [Google Scholar]
- Prakash O., Eisenberg M. A. Active transport of biotin in Escherichia coli K-12. J Bacteriol. 1974 Nov;120(2):785–791. doi: 10.1128/jb.120.2.785-791.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Rolfe B., Eisenberg M. A. Genetic and biochemical analysis of the biotin loci of Escherichia coli K-12. J Bacteriol. 1968 Aug;96(2):515–524. doi: 10.1128/jb.96.2.515-524.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe B. Lambda phage transduction of the bio A locus of Escherichia coli. Virology. 1970 Nov;42(3):643–661. doi: 10.1016/0042-6822(70)90310-7. [DOI] [PubMed] [Google Scholar]
- Vrancic A., Guha A. Evidence of two operators in the biotin locus of Escherichia coli. Nat New Biol. 1973 Sep 26;245(143):106–108. doi: 10.1038/newbio245106a0. [DOI] [PubMed] [Google Scholar]


